Back to Journals » OncoTargets and Therapy » Volume 11
High expression of USP22 predicts poor prognosis and advanced clinicopathological features in solid tumors: a meta-analysis
Authors Yang XH, Zang H, Luo Y , Wu J , Fang Z, Zhu W, Li Y
Received 9 August 2017
Accepted for publication 10 March 2018
Published 23 May 2018 Volume 2018:11 Pages 3035—3046
DOI https://doi.org/10.2147/OTT.S148662
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jianmin Xu
Xiaohui Yang,1,* Haiyang Zang,2,* Yingbin Luo,1 Jianchun Wu,1 Zhihong Fang,1 Weikang Zhu,1 Yan Li1
1Department of Oncology, Shanghai Municipal Hospital of Traditional Chinese Medicine, Shanghai University of Traditional Chinese Medicine, Shanghai, China; 2Department of Spleen and Stomach, Xinyi Municipal Hospital of Traditional Chinese Medicine, Xinyi, Jiangsu, China
*These authors contributed equally to this work
Introduction: The expression of USP22 has been demonstrated to play a pivotal role in solid tumors. However, the prognostic value of USP22 still remains unknown.
Materials and methods: A systematic meta-analysis was performed to assess the prognostic value of USP22 in cancers. A literature collection was conducted from inception to June 8, 2017 by searching PubMed, Cochrane Library, Embase, Ovid and Web of Science databases. The pooled hazard ratio (HR) and odds ratio (OR) were used to correlate high expression of USP22 with overall survival (OS) and clinicopathological features.
Results: The results, pooled by 19 studies with 2,876 cases, indicated that high expression of USP22 predicted poor OS (HR=2.48, 95% CI: 2.11–2.84, p<0.001) and disease-free survival (DFS; HR=2.55, 95% CI: 2.05–3.05, p<0.001) of cancer patients. Furthermore, high expression of USP22 was also significantly associated with advanced clinicopathological parameters, including tumor stage, tumor differentiation, metastasis, nodal status and tumor size.
Conclusion: Our finding revealed that USP22 might be an indicator of poor prognosis and advanced clinicopathological features of solid tumors and could be served as a novel biomarker.
Keywords: carcinoma, USP22, prognosis, biomarker, meta-analysis
Introduction
Carcinoma is a group of diseases involving uncontrolled cell growth with complex mechanism and unfavorable prognosis, spreading and invading other parts of the body.1 As one of the most common causes of death in the world, cancers account for approximately 13% of deaths.2 Due to inefficient diagnosis and limited treatment, the outcome of cancers tends to be poor. Thus, it is very urgent to find better prediction biomarkers to fulfill the utility and precision of diagnostic tools of carcinoma.
Deubiquitinating enzymes (DUBS), which primarily belong to the cysteine protease family, regulate several cellular mechanisms including cell cycle progression, signal transduction, growth and differentiation by catalyzing the deconjugation of ubiquitin-tagged substrates.3,4 USP22 is a subunit of DUBs with specific targets of therapeutic importance.5 As a novel deubiquitinating enzyme, USP22 plays a pivotal role in multiple physiological and pathological processes. USP22 can function as a histone-deubiquitinating component of the transcriptional regulatory histone acetylation (HAT) complex Spt-Ada-Gcn5-acetyl-transferase (SAGA).6 It removes ubiquitin efficiently from the core histones such as H2A and H2B in vitro, thereby alters the transcriptional activation and epigenetic regulation.7 Moreover, a conserved BMI-1-driven 11-gene death-from-cancer signature has been identified as a key regulator of “stemness” in cancer stem cells in recent experimental observations.8,9 Cancer stem cells characterized by the 11-gene signature are associated with metastasis, enabling anoikis resistance and aberrant cell cycle control.10 USP22 is one of the genes comprising the 11-gene polycomb/cancer stem cell signature. Besides, USP22 has been reported to be a key regulator of c-myc and p53.11,12 More importantly, depletion of USP22 results in a G1 phase cell cycle arrest in human cancer cells.13
Until now, numerous efforts have focused on the potential roles of the USP22 in various cancers. A recent study suggested that USP22 could increase TGF-β expression and promote the epithelial–mesenchymal transition (EMT).14 In addition, another study demonstrated that silencing USP22 could inhibit proliferation and induce cell cycle arrest in bladder cancer cells.15 Furthermore, USP22 was also reported with high expression level in different malignancies, such as breast cancer16 and colorectal cancer.17 Thus, these lines of evidence strongly suggested that the oncogenic role of the USP22 might contribute to progression and predict the prognosis and become an attractive therapeutic target in cancers.
However, there is no large sample report to investigate the high expression level of USP22 and prognostic value in cancers. In this study, we conducted this meta-analysis to comprehensively illustrate the prognostic role of USP22 in cancers by systematically reviewing the published evidence. We also analyzed the correlation between the expression levels of USP22 and clinicopathological features.
Materials and methods
Search strategy
The current meta-analysis was performed based on the guidelines of the preferred reporting items for systematic reviews and meta-analysis.18 A comprehensive electronic search including PubMed, Cochrane Library, Embase, Ovid and Web of Science databases was conducted to identify studies focused on the expression of USP22 in cancer patients from inception to June 8, 2017. The keywords such as “ubiquitin-specific protease 22,” “USP22” combined with “cancer,” “carcinoma,” “tumor,” “malignancy” and “neoplasm” were used to conduct our retrieval process. We also searched the references from all potential articles for any additional eligible studies. We also sent emails to the corresponding authors for missing data from articles if necessary. Other sources such as Oncomine and the Cancer Genome Altas were also searched to complete our study.
Selection criteria
The studies were enrolled in our meta-analysis if they met the following criteria: 1) studies focused on the relationship between USP22-positive expression and patients with any type of carcinoma; 2) overall survival (OS) and disease-free survival (DFS) were regarded as the end points; 3) definite testing methods were used to detect the expression level of USP22; 4) studies provided sufficient data to extract the odds ratio (OR) or hazard ratio (HR) and the corresponding 95% CI; 5) a cohort design with enough patients (not less than 40). The studies were excluded when they met the following criteria: 1) reviews, conference abstracts, editorials or case reports; 2) studies were conducted on nonhumans; 3) studies without sufficient data to calculate the HR or OR and the corresponding 95% CI; 4) studies with no information on survival outcomes. The most recent study was enrolled if multiple studies focused on the same population.
Data extraction
Data were carefully and independently extracted by two authors (XY and HZ) using a standard form based on the aforementioned selection criteria. The following information was extracted from every study: first author, publication year, ethnicity, country, sample size, age, types of tumor, median follow-up time, survival condition, testing methods, cutoff value, HR estimation and corresponding 95% CI, tumor size, TNM stage, tumor nodal metastasis, histological stage and differentiation degree. We extracted HR from multivariate analysis if both univariate and multivariate analyses were provided. Any discrepancies were resolved by consensus of all authors. We also used software Engauge Digitizer 4.1 (https://markummitchell.github.io/engauge-digitizer/) to extract data if a Kaplan–Meier curve was provided only to describe the outcome of cancer patients.
Quality assessment
The Newcastle–Ottawa Scale recommended by the Cochrane was used to assess the quality of each study enrolled in our meta-analysis.19 A star system based on selection (four stars), comparability (two stars) and outcome (three stars) was used to calculate scores. The total score ranged from 0 to 9, and studies were considered as high quality if they got 6 or more stars.
Statistical analyses
All the statistical analyses were conducted by STATA software version 12.0 (StataCorp LP, College Station, TX, USA). As for the studies focused on the prognostic value of USP22 in cancer patients, the combined HRs and the corresponding 95% CIs were used to evaluate the potential association between high expression of USP22 and OS. An HR >1 indicated poor outcome of patients with high expression of USP22. As for the pooled analysis of the association between high expression of USP22 and the clinicopathological parameters, including age, tumor stage, tumor differentiation, nodal metastasis and tumor size, the ORs with the corresponding 95% CI were used to evaluate the effects. Z-test was used to examine the pooled HRs, and the values with p<0.05 were considered statistically significant. Q-statistic test and I2 statistic test were used to measure the heterogeneity between studies. Fixed-effects model or random-effects model was performed according to the results of heterogeneity test. The random-effects model was used when the heterogeneity was considered statistically significant. Otherwise, the fixed-effects model was conducted. To estimate the accuracy when investigating the potential association between high expression of USP22 and clinical stage, we conducted a bootstrap resampling procedure to generate 1,000 resampling test to get a robust result.
Sensitivity analysis and publication bias
By omitting each study one by one, we carried out sensitivity analysis to assess the influence of every single study on the overall effect. The potential publication bias was estimated by the Begg’s funnel plot and Egger’s linear regression, with visually assessed funnel plots.
Results
Eligible studies and basic characteristics
A flowchart of the processes of identifying and selecting studies is shown in Figure 1. After a comprehensive study through the electronic database, a total of 110 studies were potentially identified for inclusion. By scanning the title and abstracts, 83 studies were excluded. Then, eight studies were excluded after full-text reading for the following reasons: four studies were reviews; two studies were nonhuman studies; one study could not provide sufficient data to extract HR and 95% CI and one study provided clinical data without survival analysis. Ultimately, 19 studies13,20–37 were enrolled in our meta-analysis.
  | Figure 1 Flowchart of the study selection process. |
The basic characteristics of the identified articles are listed in Table 1. The total sample size of all enrolled studies was 2,876 ranging from 44 to 319. All the studies were principally originated from China published from 2010 to 2016. As for the prognostic value of USP22 in various cancers, nine studies reported OS, four studies reported DFS and six studies focused on both OS and DFS. Cancer types of the patients included gastric carcinoma, esophageal squamous cell carcinoma, salivary duct carcinoma, colorectal cancer, lung adenocarcinoma, breast cancer, glioma, hepatocellular carcinoma, cervical cancer and others listed in Table 1.
Quality assessment of studies
The Newcastle–Ottawa Quality Assessment scale (NOS) was used to assess the quality of each study enrolled in our meta-analysis by two authors (YL and JW) independently. The scores of 19 enrolled studies ranged from 6 to 9, according to the evaluation criteria, which were considered as high quality. The detailed information is listed in Table S1.
Overall analysis
As for the relationship between high expression of USP22 and OS, 15 studies with 2,314 cases were enrolled in the analysis. A random-effects model was employed to evaluate the pooled HR, and the corresponding 95% CI for the heterogeneity was found between studies (I2=89.2%, p<0.001). The results of pooled analysis indicated that high expression of USP22 significantly predicted a worse OS in cancer patients (HR=2.48, 95% CI: 2.11–2.84, p<0.001; Figure 2). As for the studies evaluating DFS, 10 studies with 1,484 cases were enrolled in the analysis. The pooled result was calculated from the random-effects model for the existence of heterogeneity (I2=91.3%, p<0.001) and suggested that the elevated expression of USP22 was associated with poor DFS (HR=2.55, 95% CI: 2.05–3.05, p<0.001; Figure 3).
  | Figure 2 Association between high expression of USP22 and the OS of cancer patients. |
  | Figure 3 Association between high expression of USP22 and DFS of cancer patients. |
Meta-regression analysis
Because of the significant heterogeneity that existed between the enrolled studies, we conducted a meta-regression analysis to investigate the potential reason. We found that the publication years (t=0.42, p=0.684), sample size (t=0.10, p=0.922), median follow-up time (t=2.02, p=0.074) and biological system (t=0.56, p=0.586) could only account for minor heterogeneity for OS analysis. In addition, mean age (t=2.43, p=0.025) could account for more heterogeneity. Moreover, the results indicated that publication years (t=0.79, p=0.475), sample size (t=0.29, p=0.784), median follow-up time (t=09, p=0.934), mean age (t=0.65, p=0.553) and biological system (t=0.32, p=0.763) could explain minor heterogeneity for DFS analysis.
Subgroup analysis
We conducted subgroup analysis based on all possible factors (age, sample size, publication year, follow-up time and biological system) for evaluating both OS and DFS. When evaluating OS (as shown in Figure S1), subgroup analysis based on the age of cancer patients indicated that high expression of USP22 was significantly associated with much worse OS for patients older than 55 years (HR=2.48, 95% CI: 2.31–2.65, p<0.001) compared to the patients younger than 55 years (HR=1.82, 95% CI: 1.67–1.98, p<0.001; Figure S1A). Stratified by sample size, the data indicated a significant relationship between high expression of USP22 and cancer patients in the two groups based on sample size<150 group (HR=2.53, 95% CI: 2.35–2.73, p<0.001) and the sample size≧150 group (HR=1.92, 95% CI: 1.78–2.07, p<0.001; Figure S1B). Subgroup analysis based on the follow-up time suggested that the overexpression of USP22 predicted poor OS for both the ≧60-month group (HR=3.38, 95% CI: 2.25–4.51, p<0.001) and <60-month group (HR=2.19, 95% CI: 1.86–2.52, p<0.001; Figure S1D). As for the studies based on publication year, the results indicated that high expression of USP22 predicted poor prognosis in studies published before 2014 (HR=2.55, 95% CI: 2.06–3.04, p<0.001) and after 2014 (HR=2.41, 95% CI: 1.84–2.97, p<0.001; Figure S1C). When grouped according to different systems that the carcinoma affected, patients with carcinomas of the alimentary system had worse OS (HR=2.46, 95% CI: 2.13–2.79, p<0.001) compared to other systems (HR=2.48, 95% CI: 1.72–3.23, p<0.001; Figure S1E).
When evaluating DFS (as shown in Figure S2), subgroup analysis based on the age indicated that high expression of USP22 was significantly associated with worse DFS for patients older than 55 years (HR=2.77, 95% CI: 2.57–2.97, p<0.001) and patients younger than 55 years (HR=2.05, 95% CI: 1.82–2.28, p<0.001; Figure S2A). Being grouped according to sample size, cancer patients in the sample size <150 group (HR=2.51, 95% CI: 1.95–3.06, p<0.001) and the sample size ≧150 group (HR=2.60, 95% CI: 158–3.63, p<0.001) the data in the two groups have similar results (Figure S2B). When stratified by median follow-up time, significant results were also found in both the ≧60-month group (HR=2.98, 95% CI: 1.55–4.41, p<0.001) and <60-month group (HR=2.29, 95% CI: 1.79–2.80, p<0.001; Figure S2D). When grouped according to publication year (Figure S2C) and biological system (Figure S2D), no obvious changes occurred between groups. The studies enrolled in our analysis also contained many cancers and did not focus on any specific carcinoma. Thus, it is difficult to extend the analysis on different types of cancer. Still, we have conducted the subgroup analysis based on cancer types. We found that the overexpression of USP22 predicted worse OS in cancer patients particularly in salivary cancer (HR=3.14, 95% CI: 1.69–4.59, p=0.001), lung cancer (HR=2.46, 95% CI: 1.62–3.29, p<0.001), pancreatic cancer (HR=2.69, 95% CI: 1.70–3.69, p<0.001) and hepatocellular cancer (HR=2.13, 95% CI: 1.69–2.57, p<0.001; Figure S3A). Moreover, the results also indicated that high expression of USP22 predicted worse DFS in cancer patients particularly in salivary cancer (HR=3.09, 95% CI: 2.24–3.93, p<0.001) and colorectal cancer (HR=1.64, 95% CI: 1.33–1.95, p<0.001; Figure S3B). Furthermore, we conducted subgroup analysis based on different USP22 cutoffs. Because the detection methods when evaluating the expression of USP22 were different, the definition of high expression was also different. Thus, we redefined the high USP22 cutoffs as follows: immunohistochemistry (IHC) staining score ≥3, positive staining ≥10% and immunoreactive score≥5. The results indicated that the overexpression of USP22 predicted worse OS in cancer patients in both the high USP22 cutoff group (HR=2.52, 95% CI: 2.08–2.96, p<0.001) and the low USP22 cutoff group (HR=2.34, 95% CI: 1.99–2.68, p<0.001; Figure S4A). A similar result was also found when evaluating the association between the high level of USP22 and the DFS of cancer patients in the high USP22 cutoff group (HR=2.98, 95% CI: 2.13–3.82, p<0.001) and the low USP22 cutoff group (HR=2.02, 95% CI: 1.41–2.62, p<0.001; Figure S4B). In addition, no obvious publication bias exists because of the definition of cutoffs.
All the detailed information is summarized in Table 2.
Correlation of high expression of USP22 with clinicopathological features
To evaluate the association between high expression of USP22 and clinicopathological parameters, 15 studies were enrolled for the analysis. All the detailed data are summarized in Table 3. A fixed-effects model was used to conduct the analysis according to the heterogeneity test. The results indicated that high expression of USP22 was significantly associated with advanced tumor stage (OR=1.82, 95% CI: 1.54–2.15, p<0.001; Figure 4). We also conducted analysis assessing the clinicopathological features and expression of USP22 from tumor differentiation, metastasis, nodal status and tumor size. Pooled analysis of 15 studies suggested that the overexpression of USP22 was significantly related to tumor differentiation (poor/well+moderate; OR=1.54, 95% CI: 1.33–1.79, p<0.001; Figure S5A). Besides, 10 studies focused on the association of the expression of USP22 with cancer metastasis and suggested that high expression of USP22 was significantly associated with tumor metastasis (OR=1.54, 95% CI: 1.27–1.86, p<0.001; Figure S5B). Moreover, the results demonstrated that the overexpression of USP22 indicated a positive nodal status (OR=1.56, 95% CI: 1.34–1.82, p<0.001; Figure S5C). Furthermore, the pooled analysis showed that high expression of USP22 was significantly associated with larger tumor size (>5 cm; OR=1.25, 95% CI: 1.07–1.45, p<0.001; Figure S5D).
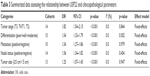  | Table 3 Summarized data assessing the relationship between USP22 and clinicopathological parameters |
  | Figure 4 Association between the overexpression of USP22 and tumor stage. |
Sensitivity analysis
By omitting one study each time, we conducted the sensitivity analysis between high expression of USP22 and survival of cancer patients to assess the stability of the results. When focused on OS, the results indicated that the impact of Zhao et al study might be more dramatic (Figure 5). However, no essential alteration was found, indicating that the results were relatively steady. Besides, when focused on DFS, the results suggested that no individual study significantly changed the pooled HRs (Figure S6).
  | Figure 5 Sensitivity analysis among studies enrolled based on the OS of cancer patients. |
Publication bias
The funnel plots used to examine publication bias of the studies on the summary of OS and DFS were relatively symmetrical, indicating that no obvious publication bias exists (OS: Figure 6; DFS: Figure S7). The results of the Egger’s test and Begg’s test also provided statistical evidence to demonstrate that no obvious publication bias was found for OS and DFS (Figures S8 and S9).
  | Figure 6 Funnel plot analysis exploring the publication bias between high expression of USP22 and OS of cancer patients. |
Resampling statistics
We applied a bootstrap resampling procedure to generate 1,000 resampling groups to evaluate the accuracy of the association between the expression of USP22 and tumor stage and obtain robust results in the current meta-analysis. Overall, ORs were analyzed for resampling statistics. The results (pooled OR=2.53, 95% CI: 2.47–2.59) indicated that our result was relatively robust, which is presented in Supplementary Excel file.
Discussion
Despite significant advances in supervision and clinical treatment strategies in recent decades, the overall prognosis of cancers still remains dismal.38 The existing TNM stage might be the main method to predict the outcome of cancer patients; however, it is not timely enough to begin initiate intervention for treatment.39 It has to be solved urgently to classify the patients with a high possibility of tumor recurrence and determine the probable outcome. Thus, it is an essential step to identify effective biomarkers associated with metastasis and survival to improve the prognosis of cancer patients.
Recently, cancer epigenetics is one of the main fields of many of the most significant and exciting advances in cancer study.40 Glinsky9 proposed that 11-gene polycomb/cancer stem cell signature could be used as an effective predictor to assess the therapeutic outcome of individual cancer patients. As one pivotal role of this signature, USP22 has been demonstrated to function as a highly promising diagnostic marker in the pathological process of epithelial malignancies and other solid tumors.23,41 The targeted deubiquitylation of histones is predominantly associated with transcriptional activation and cancer progression.42 USP22, as part of the mammalian SAGA (Spt-Ada-Gcn5) complex, edits the histone code by deubiquitinating H2A and H2B.43 Within the past few years, numerous studies have explored the potential relationship between the expression of USP22 and the survival outcome of cancer patients.1,24 However, the results still remain ambiguous. In the current study, we conducted a comprehensive meta-analysis to explore the exact role of USP22 in cancers. This might be the first meta-analysis evaluating the association between USP22 and the prognosis and clinicopathological parameters of cancers.
Our data indicated that the overexpression of USP22 was significantly associated with the survival of cancer patients. High expression of USP22 could predict poor OS and DFS for cancers (p<0.001 and p<0.001, respectively). Because of the high heterogeneity existing in pooled effects, we conducted a meta-regression analysis to investigate the potential reason. The results indicated that publication year, sample size, median follow-up time, mean age and biological system could explain minor heterogeneity for OS and DFS analysis. Despite that, we still performed subgroup analysis based on publication year, sample size, median follow-up time, mean age and biological system. The results suggested that high expression of USP22 predicted worse OS for older patients (≧55, HR=2.48, 95% CI: 2.31–2.65, p<0.001) compared to relatively younger patients. In patients with cancer belonging to alimentary system compared to other system, the high expression of USP22 predicted worse OS (HR=2.48, 95% CI: 1.72–3.23, p<0.001). In addition, no obvious alteration occurred when stratified by publication year, follow-up time and sample size.
There are some clues that might help to explain why USP22 could predict the overall survival of cancer patients. Some scholars presented that deubiquitylation of USP22 could regulate transcriptional activation.44 As a critical fraction of malignant transformation in mammalian cells, the Myc-driven transcription depends on hSAGA recruitment, which contains USP22.45 What is more is that Myc oncoprotein could recruit USP22 to specific target gene loci and other sequence-specific activators.6 In addition, it is reported that USP22 recruitment could transcriptionally activate certain essential cell cycle genes, which resulted in a G1 phase cell cycle arrest.46
To further assess the prognostic role of USP22, we also linked it to clinicopathological features of cancers. The data indicated that high expression of USP22 was significantly associated with advanced tumor stage (OR=1.82, 95% CI: 1.54–2.15, p<0.001). The overexpression of USP22 was also significantly associated with poor tumor differentiation, distant tumor metastasis, positive nodal status and larger tumor size (>5 cm). All the results showed that USP22 predicted worse clinicopathological features, which was consistent with the OS analysis.
However, some limitations still exist in our analysis. First, the determination of high expression of USP22 might differ in the studies for the different cutoff values, which may cause potential bias. Second, the calculation errors were unavoidable because Engage Digitizer 4.1 was used to estimate the data when only Kaplan–Meier curve was provided. Third, the sample size enrolled in our analysis was relatively small. Fourth, large heterogeneity still exists in our study despite the random-effects model being used to conduct the analysis. Finally, the literature was limited to English, which might exclude potential studies published in other languages.
Conclusion
The current meta-analysis demonstrated that the overexpression of USP22 was significantly associated with poor OS and DFS of cancer patients. Furthermore, the high level of expression of USP22 was highly correlated with advanced clinicopathological features. The investigation of USP22 might be a reasonable approach to predict cancer patient prognosis and restrain cancer progression. Due to the limitation of the study, further high-quality studies with a large sample are encouraged to fulfill our results.
Acknowledgments
This study was supported by Shanghai Municipal Health Authority’s major project (20134016), Science and Technology Commission Shanghai Municipalit’s major project (17401933500), and Natural Science Foundation of China (81673947).
Disclosure
The authors report no conflicts of interest in this work.
References
Coleman MP. Cancer survival: global surveillance will stimulate health policy and improve equity. Lancet. 2014;383(9916):564–573. | ||
Allemani C, Weir HK, Carreira H, et al. Global surveillance of cancer survival 1995–2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet. 2015;385(9972):977–1010. | ||
Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. | ||
Hoeller D, Dikic I. Targeting the ubiquitin system in cancer therapy. Nature. 2009;458(7237):438–444. | ||
Love KR, Catic A, Schlieker C, Ploegh HL. Mechanisms, biology and inhibitors of deubiquitinating enzymes. Nat Chem Biol. 2007;3(11):697–705. | ||
Zhang XY, Varthi M, Sykes SM, et al. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol Cell. 2008;29(1):102–111. | ||
Armour SM, Bennett EJ, Braun CR, et al. A high-confidence interaction map identifies SIRT1 as a mediator of acetylation of USP22 and the SAGA coactivator complex. Mol Cell Biol. 2013;33(8):1487–1502. | ||
Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest. 2005;115(6):1503–1521. | ||
Glinsky GV. “Stemness” genomics law governs clinical behavior of human cancer: implications for decision making in disease management. J Clin Oncol. 2008;26(17):2846–2853. | ||
Glinsky GV. Genomic models of metastatic cancer: functional analysis of death-from-cancer signature genes reveals aneuploid, anoikis-resistant, metastasis-enabling phenotype with altered cell cycle control and activated Polycomb Group (PcG) protein chromatin silencing pathway. Cell Cycle. 2006;5(11):1208–1216. | ||
Liu Y, Yang Y, Xu H, Dong X. Implication of USP22 in the regulation of BMI-1, c-Myc, p16INK4a, p14ARF, and cyclin D2 expression in primary colorectal carcinomas. Diagn Mol Pathol. 2010;19(4):194–200. | ||
Lin Z, Yang H, Kong Q, et al. USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Mol Cell. 2012;46(4):484–494. | ||
Ning Z, Wang A, Liang J, et al. USP22 promotes the G1/S phase transition by upregulating FoxM1 expression via beta-catenin nuclear localization and is associated with poor prognosis in stage II pancreatic ductal adenocarcinoma. Int J Oncol. 2014;45(4):1594–1608. | ||
Zhang J, Luo N, Tian Y, et al. USP22 knockdown enhanced chemosensitivity of hepatocellular carcinoma cells to 5-Fu by up-regulation of Smad4 and suppression of Akt. Oncotarget. 2017;8(15):24728–24740. | ||
Lv L, Xiao XY, Gu ZH, Zeng FQ, Huang LQ, Jiang GS. Silencing USP22 by asymmetric structure of interfering RNA inhibits proliferation and induces cell cycle arrest in bladder cancer cells. Mol Cell Biochem. 2011;346(1–2):11–21. | ||
Kim D, Hong A, Park HI, et al. Deubiquitinating enzyme USP22 positively regulates c-Myc stability and tumorigenic activity in mammalian and breast cancer cells. J Cell Physiol. 2017;232(12):3664–3676. | ||
Li Y, Yang Y, Li J, et al. USP22 drives colorectal cancer invasion and metastasis via epithelial-mesenchymal transition by activating AP4. Oncotarget. 2017;8(20):32683–32695. | ||
Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. J Clin Epidemiol. 2009;62(10):e1–e34. | ||
Oremus M, Oremus C, Hall GB, McKinnon MC; ECT & Cognition Systematic Review Team. Inter-rater and test-retest reliability of quality assessments by novice student raters using the Jadad and Newcastle-Ottawa Scales. BMJ Open. 2012;2(4):pii:e001368. | ||
Li J, Wang Z, Li Y. USP22 nuclear expression is significantly associated with progression and unfavorable clinical outcome in human esophageal squamous cell carcinoma. J Cancer Res Clin Oncol. 2012;138(8):1291–1297. | ||
Piao S, Ma J, Wang W, et al. Increased expression of USP22 is associated with disease progression and patient prognosis of salivary duct carcinoma. Oral Oncol. 2013;49(8):796–801. | ||
Yang DD, Cui BB, Sun LY, et al. The co-expression of USP22 and BMI-1 may promote cancer progression and predict therapy failure in gastric carcinoma. Cell Biochem Biophys. 2011;61(3):703–710. | ||
Liu YL, Yang YM, Xu H, Dong XS. Aberrant expression of USP22 is associated with liver metastasis and poor prognosis of colorectal cancer. J Surg Oncol. 2011;103(3):283–289. | ||
Hu J, Yang D, Zhang H, et al. USP22 promotes tumor progression and induces epithelial-mesenchymal transition in lung adenocarcinoma. Lung Cancer. 2015;88(3):239–245. | ||
Wang Z, Zhu L, Guo T, Wang Y, Yang J. Decreased H2B monoubiquitination and overexpression of ubiquitin-specific protease enzyme 22 in malignant colon carcinoma. Hum Pathol. 2015;46(7):1006–1014. | ||
Yang M, Liu YD, Wang YY, Liu TB, Ge TT, Lou G. Ubiquitin-specific protease 22: a novel molecular biomarker in cervical cancer prognosis and therapeutics. Tumour Biol. 2014;35(2):929–934. | ||
Zhang Y, Yao L, Zhang X, et al. Elevated expression of USP22 in correlation with poor prognosis in patients with invasive breast cancer. J Cancer Res Clin Oncol. 2011;137(8):1245–1253. | ||
Liang J, Zhang X, Xie S, et al. Ubiquitin-specific protease 22: a novel molecular biomarker in glioma prognosis and therapeutics. Med Oncol. 2014;31(4):899. | ||
Wang H, Li YP, Chen JH, et al. Prognostic significance of USP22 as an oncogene in papillary thyroid carcinoma. Tumour Biol. 2013;34(3):1635–1639. | ||
Ning J, Zhang J, Liu W, Lang Y, Xue Y, Xu S. Overexpression of ubiquitin-specific protease 22 predicts poor survival in patients with early-stage non-small cell lung cancer. Eur J Histochem. 2012;56(4):e46. | ||
Tang B, Liang X, Tang F, et al. Expression of USP22 and Survivin is an indicator of malignant behavior in hepatocellular carcinoma. Int J Oncol. 2015;47(6):2208–2216. | ||
Liu YL, Yang YM, Xu H, Dong XS. Increased expression of ubiquitin-specific protease 22 can promote cancer progression and predict therapy failure in human colorectal cancer. J Gastroenterol Hepatol. 2010;25(11):1800–1805. | ||
Tang B, Tang F, Li B, et al. High USP22 expression indicates poor prognosis in hepatocellular carcinoma. Oncotarget. 2015;6(14):12654–12667. | ||
Zhao HD, Tang HL, Liu NN, et al. Targeting ubiquitin-specific protease 22 suppresses growth and metastasis of anaplastic thyroid carcinoma. Oncotarget. 2016;7(21):31191–31203. | ||
Liang JX, Ning Z, Gao W, et al. Ubiquitin-specific protease 22 induced autophagy is correlated with poor prognosis of pancreatic cancer. Oncol Rep. 2014;32(6):2726–2734. | ||
Piao S, Liu Y, Hu J, et al. USP22 is useful as a novel molecular marker for predicting disease progression and patient prognosis of oral squamous cell carcinoma. PLoS One. 2012;7(8):e42540. | ||
Dai W, Yao Y, Zhou Q, Sun CF. Ubiquitin-specific peptidase 22, a histone deubiquitinating enzyme, is a novel poor prognostic factor for salivary adenoid cystic carcinoma. PLoS One. 2014;9(1):e87148. | ||
Karapanagiotou EM, Terpos E, Dilana KD, et al. Serum bone turnover markers may be involved in the metastatic potential of lung cancer patients. Med Oncol. 2010;27(2):332–338. | ||
Le QT, Chen E, Salim A, et al. An evaluation of tumor oxygenation and gene expression in patients with early stage non-small cell lung cancers. Clin Cancer Res. 2006;12(5):1507–1514. | ||
Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4(2):143–153. | ||
Atanassov BS, Dent SY. USP22 regulates cell proliferation by deubiquitinating the transcriptional regulator FBP1. EMBO Rep. 2011;12(9):924–930. | ||
Pijnappel WW, Timmers HT. Dubbing SAGA unveils new epigenetic crosstalk. Mol Cell. 2008;29(2):152–154. | ||
Chipumuro E, Henriksen MA. The ubiquitin hydrolase USP22 contributes to 3′-end processing of JAK-STAT-inducible genes. FASEB J. 2012;26(2):842–854. | ||
Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447(7143):407–412. | ||
Bouchard C, Dittrich O, Kiermaier A, et al. Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes Dev. 2001;15(16):2042–2047. | ||
Guney I, Wu S, Sedivy JM. Reduced c-Myc signaling triggers telomere-independent senescence by regulating Bmi-1 and p16(INK4a). Proc Natl Acad Sci U S A. 2006;103(10):3645–3650. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


