Back to Journals » OncoTargets and Therapy » Volume 8
High DEPTOR expression correlates with poor prognosis in patients with esophageal squamous cell carcinoma
Authors Liu N, Zhang J, Liu Y, Li J, Zhang Z, Li J, Liu W, Huang C, Shen T, Gu C, Gao D, Wu X, Wu X, Liu D, Gao M, Jiang L, Kang H
Received 22 July 2015
Accepted for publication 25 September 2015
Published 19 November 2015 Volume 2015:8 Pages 3449—3455
DOI https://doi.org/10.2147/OTT.S92862
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Faris Farassati
Nan-bo Liu,1,* Jun-hua Zhang,2,* Yu-fan Liu,1,* Jun Li,3,* Zhen-zhong Zhang,1 Ji-wei Li,1 Wen-yue Liu,1 Chen Huang,1,4 Tao Shen,5 Cheng-wei Gu,6 Dong-yun Gao,7 Xia Wu,8 Xu Wu1
1Department of Thoracic Surgery, 2Department of Anesthesiology, Nanfang Hospital, Southern Medical University, 3Department of Thoracic Surgery, The Third Affiliated Hospital of Southern Medical University, Guangzhou, 4Department of Thoracic Surgery, Fujian Provincial Hospital, Fuzhou, 5Department of Thoracic Surgery, Jiangmen Central Hospital, Jiangmen, 6Department of Thoracic Surgery, The First Affiliated Hospital of Xinxiang Medical University, Xinxiang, 7Department of Oncology, Dongtai People’s Hospital, Dongtai, 8Department of Breast Cancer, Affiliated Hospital, Academy of Military Medical Sciences, Beijing, People’s Republic of China
*These authors contributed equally to this work
Objective: The disheveled, Egl-10, and pleckstrin (DEP) domain containing mammalian target of rapamycin (mTOR)-interacting protein (DEPTOR) is a binding protein containing mTOR complex 1 (mTORC1), mTOR complex 2 (mTORC2), and an endogenous mTOR inhibitor. DEPTOR shows abnormal expressions in numerous types of solid tumors. However, how DEPTOR is expressed in esophageal squamous cell carcinoma (ESCC) remains elusive.
Methods: The expression of DEPTOR in 220 cases of ESCC and non-cancerous adjacent tissues was detected by immunohistochemistry. DEPTOR levels in ESCC and paired normal tissue were quantified using reverse transcription-polymerase chain reaction and Western blot analysis to verify the immunohistochemical results. The relationship between DEPTOR expression and the clinicopathological features of ESCC was analyzed based on the results of immunohistochemistry. Finally, we analyzed the relationship between DEPTOR expression and the prognosis of patients with ESCC.
Results: Immunohistochemical staining showed that the expression rate of DEPTOR in ESCC tissues was significantly increased. DEPTOR mRNA and protein expression was significantly higher in ESCC tissues than in normal adjacent esophageal squamous tissues. High DEPTOR expression was significantly correlated with regional lymph node status in the TNM stage of patients with ESCC. Kaplan–Meier survival curves showed that the rate of overall survival was significantly lower in patients with high DEPTOR expression than in those with low DEPTOR expression. Additionally, high DEPTOR expression was an independent prognostic predictor for ESCC patients.
Conclusion: High DEPTOR expression is an independent prognostic biomarker indicating a worse prognosis for patients with ESCC.
Keywords: DEPTOR, ESCC, prognosis
Introduction
Esophagectomy remains the primary treatment for patients with esophageal squamous cell carcinoma (ESCC). Although the survival rates have improved over the past decade, the 5-year survival rate is still only 30%.1 Most tumor recurrences occur within the first year after surgery and are typically fatal. However, patients surviving for 5 years have approximately the same life expectancy as the general population.1,2 The postoperative recurrence, invasion, and metastasis of ESCC are affected by many closely linked factors such as proto-oncogene activation, tumor-suppressor gene inactivation, and abnormal protein expression.
Previous studies have shown that the mammalian target of rapamycin (mTOR) channel plays an important role in the regulation of protein synthesis and is activated in many human cancers.3,4 Therefore, the mTOR inhibitor, a rapamycin derivative, is used to inhibit the transcription of cancer gene(s), the growth of cancer, and the generation of the tumor vasculature caused by abnormal signal channels, and has been examined in recent studies.5–7 Some scholars have suggested that a coherent understanding of the mTOR network is imperative, as its dysregulation has been implicated in diverse pathologies including cancer, diabetes, autism, and aging.8 In addition, a study has shown that disheveled, Egl-10, and pleckstrin (DEP) domain containing mammalian target of rapamycin (mTOR)-interacting protein (DEPTOR) abnormal expression may be associated with resistance to chemotherapy in breast cancer patients; these authors also highlighted the dichotomous functions of DEPTOR in modulating the proliferation and survival of triple-negative breast cancers during metastasis.9 DEPTOR is a binding protein containing mTOR complex 1(mTORC1), mTOR complex 2 (mTORC2), and an endogenous mTOR inhibitor. DEPTOR shows abnormal expression in numerous types of solid tumors.2 However, how DEPTOR is expressed in ESCC remains unclear.
Materials and methods
Patients and specimens
ESCC was diagnosed in patients, who were enrolled in the current study, at the Nanfang Hospital and The Third Affiliated Hospital of the Southern Medical University, Guangzhou, People’s Republic of China from 2004 to 2008. Patients included in the current study had undergone esophageal cancer resection. Pathological review confirmed ESCC in resected tumors and carcinoma-adjacent non-tumorous esophageal tissues. All samples were obtained from the two participating institutions, and were identified using anonymous codes that met the requirements of Nanfang Hospital of Southern Medical University and The Third Affiliated Hospital of Southern Medical University’s guidelines, as formulated based on the Declaration of Helsinki. Patients were required to have provided written informed consent, and the Review Board of the Southern Medical University approved the protocol. The American Joint Committee on Cancer/Union for International Cancer Control (AJCC/UICC) classification guidelines were used as criteria to divide the ESCC specimens into different phases, while World Health Organization (WHO) criteria were applied for subtyping the grading and histopathology of ESCC specimens. ESCC biopsy samples and matching non-cancerous esophageal tissues were frozen in liquid nitrogen and stored.
Immunohistochemistry
Immunohistochemical analysis using streptomycin avidin-biotin peroxidase complex (SP) method was conducted to evaluate paraffin sections. The immunohistochemical staining procedure was conducted according to the manufacturer’s instructions, mainly including paraffin sectioning, regular dewaxing to water, antigen retrieval, 3% hydrogen peroxide blocking of endogenous peroxidase, goat serum closing, rabbit polyclonal antibody against human DEPTOR (1:200; BS-8255R; Beijing Biosynthesis Biotechnology Co, Ltd, Beijing, People’s Republic of China), secondary antibodies, SP, 3,3′-Diaminobenzidine (DAB) staining, and a contrast dye seal. Biopsies of known normal tissues were used as positive controls, while phosphate-buffered saline (PBS) was substituted for primary antibodies in the negative controls.
A single-blind study was conducted to read the biopsies (pathologists were blinded to the sample information). Five medium views were randomly selected (200 times), and 200 cancer cells were counted in every view; thus, a total of 1,000 cells were counted and were divided into the following types according to the staining intensities: score 0, no staining; score 1, light yellow staining; score 2, brown yellow staining; and score 3, sepia staining. Samples were classified as 0%–100% according to the proportion of cells with positive cell nuclei. In the present study, the product >0 from the staining intensity multiplied by the positive proportion was defined as DEPTOR-positive; otherwise, the sample was considered to be negative. In addition, DEPTOR staining intensities with scores of 0 and 1 were defined as having low expression of DEPTOR, while those with scores of 2 and 3 were defined as having high expression.
Reverse transcription-polymerase chain reaction
Trizol reagent was used to extract total RNA from the cells, and absorbance was measured at 260 nm and 280 nm. RNA content was also determined. Photos were taken after 1% Sepharose gel electrophoresis. Reverse transcription of the mRNA was conducted according to the reverse transcription-polymerase chain reaction (RT-PCR) guidance manual to produce cDNA, which was then used as a template for PCR amplification for 30 cycles under the following PCR conditions: 94°C for 5 minutes; 94°C for 30 seconds; 56°C for 30 seconds; and 72°C for 40 seconds. Final extension was conducted at 72°C for 5 minutes. The primer sequences of DEPTOR were as follows: forward, 5′-TTTGTGGTGCGAGGAAGTAA-3′; reverse, 5′-CATTGCTTTGTGTCATTCTGG-3′. The amplified products were separated by electrophoresis, and the gel image was photographed under ultraviolet light. Quantity One® software was used to analyze the electrophoresis banding gray values with glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as the standardized internal reference.
Western blot analysis
Protein was extracted from the tissue lysate, and the bicinchoninic acid assay (BCA) method was used to measure protein concentration. Proteins were separated by 15% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), were transferred to a membrane, and were blocked with 5% skim milk for 1 hour. DEPTOR and GAPDH primary antibodies were incubated with the membranes overnight, after which the membranes were washed, and immunoglobulin G (IgG)/biotin IgG secondary antibodies were incubated with the membrane for 2 hours. After incubation, the membrane was washed three times with mixture of Tris-buffered saline and Tween 20 (TBST). Finally, the electrogenerated chemiluminescence (ECL) method was used to detect the protein bands. A Gel-Pro analyzer was used to analyze band absorbance, with GAPDH used as the standardized internal reference.
Statistical methods
To study the relationship between DEPTOR expression and patient characteristics including age, sex, tumor stage, TNM classification, histological differentiation, and vital status, we applied Pearson’s chi-square test and Spearman’s correlation analysis. The Kaplan–Meier method was used to draw the survival curves for patients with both DEPTOR-high and DEPTOR-low expression, while the log-rank test was used to compare statistical differences. Overall survival (OS) was calculated from the date of surgery to the date of death or last follow-up. Cox regression analysis was conducted to analyze univariable and multivariable survival. P-values <0.05 were regarded as statistically significant. SPSS version 13.0 (SPSS Inc, Chicago, IL, USA) software was used for statistical analyses.
Results
DEPTOR expression in ESCC and adjacent tissues
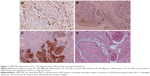
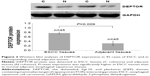
DEPTOR expression was detected in 220 cases of ESCC and in corresponding adjacent tissue samples by immunohistochemistry. Immunohistochemical staining showed that DEPTOR was predominantly localized in the nucleus or cytoplasm of cancer and normal esophageal squamous cells (Figure 1). Compared with normal adjacent esophageal squamous tissues, the high expression rate of DEPTOR in ESCC tissues was significantly increased (P=0.002). To confirm this finding, we investigated DEPTOR expression in 45 cases of ESCC and in corresponding normal adjacent tissues using RT-PCR and Western blot analysis. The results indicated that DEPTOR mRNA and protein expression was significantly higher in ESCC tissues than in normal adjacent esophageal squamous tissues (P=0.0006 and P=0.009, respectively; Figures 2 and 3, respectively).
DEPTOR expression and clinicopathological characteristics of ESCC
We analyzed the relationship between the expression of DEPTOR and clinicopathological factors of patients with ESCC based on immunohistochemical results. High DEPTOR expression was significantly correlated with regional lymph node status in the TNM stage of patients (P<0.05). However, high DEPTOR expression was not significantly correlated with other clinical parameters including age, sex, tumor size, degree of differentiation, location, smoking history, and radio-chemotherapy (P>0.05) (Table 1).
DEPTOR-high expression as a prognostic marker for ESCC
In the current study, the primary outcome was the OS of patients. In order to investigate the relationship between DEPTOR expression and patient prognosis, we plotted OS curves using the Kaplan–Meier method. Kaplan–Meier survival curves showed that the rate of OS was significantly lower in patients with high DEPTOR expression than in those with low DEPTOR expression (P<0.001; Figure 4). Subgroup analysis showed the same results (Figure 4). This indicates that patients with high DEPTOR expression had significantly shorter OS than those with low expression. We performed univariate and multivariate analysis using the Cox regression model. In addition to lymph node metastasis and TNM stage, high DEPTOR expression was also an independent prognostic predictor for ESCC patients (Table 2).
Discussion
mTOR is a protein kinase that interacts with rapamycin in mammalian cells.10 It is composed of 2,549 amino acids and has a relative molecular mass of 289 KDa.11 mTOR consists of two isoforms, mTORC1 and mTORC2, which have different functions because of their different subunits.12 DEPTOR is an important constituent of both of these isoforms.13 The mTOR signal transduction pathway can inhibit cancer cell apoptosis, maintain cell survival, and promote tumor neovascularization.7,14 It plays an important role in the generation and metastasis of malignant tumors, and thus is a well-studied signaling pathway for targeted therapy. For example, both Torisel® and everolimus are derivatives of rapamycin and can reduce the activity of mTOR while working in conjunction with a drug that prohibits the proliferation of breast cancer cells. This treatment is viable for patients with advanced and/or metastatic breast cancer.15
However, some studies have shown that DEPTOR was highly expressed in some solid tumors, possibly because DEPTOR not only inhibited the activity of mTORC1, but also reduced the negative feedback loop that inhibits the function of mTORC1 on phosphatidylinositol 3 kinase (P13K) through inhibiting ribosomal protein S6 kinase 1 (S6K1) to activate protein kinase B (Akt).16–18 Activated Akt factors may regulate many molecular families involved in cell apoptosis, inhibit tumor cell apoptosis, and disrupt the balance between cell proliferation and apoptosis in tissues to maintain cell survival.19 Moreover, active P13K and Akt may also upregulate HIF1d through other channels to promote the expression of vascular endothelial growth factor, which stimulates the generation of tumor vessels, thus providing sufficient nutrition to the tumor.17,20 However, the inhibition of DEPTOR expression may also promote apoptosis. Therefore, overexpression of DEPTOR may promote the generation and development of some malignant tumors. However, the role of DEPTOR in esophageal cancer or the relationship between its expression and the generation, development, and prognosis of esophageal cancer remains unclear.
In the present study, DEPTOR was identified in ESCC tissues and normal esophageal mucosa by immunohistochemistry. We found that the rate of DEPTOR-high expression was significantly higher in ESCC tissues than in corresponding adjacent tissues. To verify this result, DEPTOR was detected on the mRNA level by RT-PCR analysis. The mRNA levels of DEPTOR in ESCC tissues were significantly higher than those in normal esophageal tissues. Additionally, we investigated the relationship between DEPTOR expression levels and clinicopathological parameters of ESCC patients. The results showed that DEPTOR expression was significantly correlated with lymph node status and TNM stage, but not with age, sex, differentiation, location, and T stage of patients with ESCC. Through long-term follow-up, the survival rates of the ESCC patients with high DEPTOR expression were significantly lower than in patients with low DEPTOR expression. Univariate analysis showed that DEPTOR expression, lymph node status, and TNM stage were correlated with the OS of patients with ESCC. Multivariate analysis further revealed that high DEPTOR expression independently predicted unfavorable OS of ESCC patients.
Conclusion
In conclusion, high DEPTOR expression is an independent prognostic biomarker that indicates a worse prognosis for patients with ESCC, and the specific mechanisms require further analysis.
Acknowledgment
This work was supported by the Foundation of the President of Nanfang Hospital, Southern Medical University, Guangzhou, People’s Republic of China (grant 2013B016).
Disclosure
The authors report no conflicts of interest in this work.
References
Rouvelas I, Zeng W, Lindblad M, Viklund P, Ye W, Lagergren J. Survival after surgery for oesophageal cancer: a population-based study. Lancet Oncol. 2005;6(11):864–870. | ||
Derogar M, Lagergren P. Health-related quality of life among 5-year survivors of esophageal cancer surgery: a prospective population-based study. J Clin Oncol. 2012;30(4):413–418. | ||
Yang H, Rudge DG, Koos JD, Vaidialingam B, Yang HJ, Pavletich NP. mTOR kinase structure, mechanism and regulation. Nature. 2013;497(7448):217–223. | ||
Souroullas GP, Sharpless NE. mTOR signaling in melanoma: oncogene-induced pseudo- senescence? Cancer Cell. 2015;27(1):3–5. | ||
Northrup H. The mTOR inhibitor revolution rolls on. Lancet Oncol. 2014;15(13):1418–1419. | ||
Chawla SP, Staddon AP, Baker LH, et al. Phase II study of the mammalian target of rapamycin inhibitor ridaforolimus in patients with advanced bone and soft tissue sarcomas. J Clin Oncol. 2012;30(1):78–84. | ||
Wang Z, Zhong J, Inuzuka H, et al. An evolving role for DEPTOR in tumor development and progression. Neoplasia. 2012;14(5):368–375. | ||
Dibble CC, Cantley LC. Regulation of mTORC1 by PI3K signaling. Trends Cell Biol. 2015;25(9):545–555. | ||
Parvani JG, Davuluri G, Wendt MK, et al. Deptor enhances triple-negative breast cancer metastasis and chemoresistance through coupling to survivin expression. Neoplasia. 2015;17(3):317–328. | ||
Gentzler RD, Altman JK, Platanias LC. An overview of the mTOR pathway as a target in cancer therapy. Expert Opin Ther Targets. 2012;16(5):481–489. | ||
Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. | ||
Altman JK, Szilard A, Goussetis DJ, et al. Autophagy is a survival mechanism of acute myelogenous leukemia precursors during dual mTORC2/mTORC1 targeting. Clin Cancer Res. 2014;20(9):2400–2409. | ||
Yang Y, Bardeleben C, Frost P, et al. DEPTOR is linked to a TORC1-p21 survival proliferation pathway in multiple myeloma cells. Genes Cancer. 2014;(11–12):407–419. | ||
Bruneau S, Nakayama H, Woda CB, Flynn EA, Briscoe DM. DEPTOR regulates vascular endothelial cell activation and proinflammatory and angiogenic responses. Blood. 2013;122(10):1833–1842. | ||
Zagouri F, Sergentanis TN, Chrysikos D, Filipits M, Bartsch R. mTOR inhibitors in breast cancer: a systematic review. Gynecol Oncol. 2012;127(3):662–672. | ||
Li H, Sun GY, Zhao Y, et al. DEPTOR has growth suppression activity against pancreatic cancer cells. Oncotarget. 2014;5(24):12811–12819. | ||
Zhang HR, Chen JM, Zeng ZY, Que WZ. Knockdown of DEPTOR inhibits cell proliferation and increases chemosensitivity to melphalan in human multiple myeloma RPMI-8226 cells via inhibiting PI3K/AKT activity. J Int Med Res. 2013;41(3):584–595. | ||
Corominas-Faja B, Cufí S, Oliveras-Ferraros C, et al. Nuclear reprogramming of luminal-like breast cancer cells generates Sox2-overexpressing cancer stem-like cellular states harboring transcriptional activation of the mTOR pathway. Cell Cycle. 2013;12(18):3109–3124. | ||
Ma WW, Adjei AA. Novel agents on the horizon for cancer therapy. CA Cancer J Clin. 2009;59(2):111–137. | ||
Pande M, Bondy ML, Do KA, et al. Association between germline single nucleotide polymorphisms in the PI3K-AKT-mTOR pathway, obesity, and breast cancer disease-free survival. Breast Cancer Res Treat. 2014;147(2):381–387. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.