Back to Journals » OncoTargets and Therapy » Volume 8
GIT1 is a novel prognostic biomarker and facilitates tumor progression via activating ERK/MMP9 signaling in hepatocellular carcinoma
Authors Chen J, Yang P, Yang J, Wen Z, Zhang B, Zheng X
Received 19 September 2015
Accepted for publication 12 November 2015
Published 15 December 2015 Volume 2015:8 Pages 3731—3742
DOI https://doi.org/10.2147/OTT.S96715
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Daniele Santini
Junyi Chen,1,* Pinghua Yang,2,* Jue Yang,2 Zhijian Wen,2 Baohua Zhang,2 Xin Zheng1,3
1Department of General Surgery, Branch of the First People’s Hospital of Shanghai, 2Department of Hepatic Surgery, the Eastern Hepatobiliary Surgery Hospital, Second Military Medical University, 3Department of Traditional Chinese Medicine, Branch of the first People’s Hospital of Shanghai, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Aim: Multiple studies have revealed that G-protein-coupled receptor kinase-interacting protein 1 (GIT1) is overexpressed in many cancers and facilitates tumor progression. However, the role of GIT1 in hepatocellular carcinoma (HCC) remains unclear.
Methods: GIT1 expression was detected in cell lines and 130 pairs of HCC and matched adjacent noncancerous samples. Transwell assay, flow cytometry, caspase 3/7 activity assay, 5-bromodeoxyuridine cell proliferation assay, and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay were used to assess invasion, migration, apoptosis, and proliferation of HCC cells. Furthermore, GIT1 expression was detected by immunohistochemistry to evaluate its correlation with phospho-extracellular signal-regulated kinase (p-ERK)1/2. The regulatory effect of GIT1 on ERK1/2, p-ERK1/2, and matrix metalloproteinase-9 (MMP9) in HCC cells was confirmed by immunoblotting.
Results: In this study, we demonstrated that GIT1 was more highly expressed in HCC samples than that in non-HCC samples, and overexpression of GIT1 was correlated with clinicopathological features of poor prognosis. Clinical analysis demonstrated that GIT1 is an independent prognostic biomarker for predicting overall survival and disease-free survival of patients with HCC. In vitro studies showed that downregulation of GIT1 facilitated HCC cell apoptosis and repressed HCC cell invasion, migration, and proliferation. Overexpression of GIT1 is associated with p-ERK1/2 amplification in HCC tissues. Moreover, downregulation of GIT1 resulted in inactivation of ERK signaling and downregulation of MMP9.
Conclusion: Our findings indicate that GIT1 is an independent prognostic biomarker and facilitates HCC progression via activating ERK/MMP9 signaling.
Keywords: GIT1, HCC, prognosis, ERK signaling
Introduction
Hepatocellular carcinoma (HCC) is the fifth most frequent cancer in the world and the second major cause of cancer-related death in the People’s Republic of China.1,2 The incidence of HCC is increasing in many western countries due to the rising morbidity of hepatitis C virus infection.3,4 Tumor recurrence and metastases contribute predominantly to the high mortality rate of HCC patients after hepatic resection.5–7 Despite many advances in the treatment of HCC such as surgical resection, chemotherapy, radiofrequency ablation, and transarterial therapy, the prognosis of patients with HCC remains very poor.8,9 Therefore, it is critical to understand the mechanisms involved in hepatocarcinogenesis and explore prognostic biomarkers and therapeutic targets of HCC.
G-protein-coupled receptor kinase-interacting protein 1 (GIT1) was originally considered as a multifunctional scaffold protein, and was suggested to be associated with paxillin and capable of binding and regulating various proteins.10,11 As a ubiquitously expressed scaffold protein, GIT1 has emerged as an important transducer of variety of extracellular signals.12,13 Notably, GIT1 is known to play an important role in promoting focal adhesion formation and cell migration.14–16 Furthermore, several studies suggested an overexpression of GIT1 in carcinogenesis of multiple tumor types.14,17,18 High-expression level of GIT1 correlates with cancer invasion and migration in melanoma.11,19 Importantly, overexpression of GIT1-induced robust extracellular signal-regulated kinase (ERK) signaling activation has been found in multiple cancer types.20–22 However, the exact role and molecular mechanisms involved in GIT1 are still unclear in HCC.
ERK signaling pathway is very important in facilitating tumor cell invasion, migration, and proliferation, and is often dysregulated in many cancers.23,24 It has been reported that ERK signaling plays a significant role in promoting HCC initiation and progression.25 Notably, while ERK signaling is understood to a broader extent in multiple tumor types, new evidence has emerged revealing that ERK activation can occur in GIT1-dependent pattern.20 Multiple studies have already explored a novel molecular mechanism of GIT1-mediated ERK activation, which leads to the initiation and progression of multiple tumor types.21,22 Furthermore, it was recently reported that matrix metalloproteinase-9 (MMP9) expression is regulated by the ERK signaling in human cancers.26,27 In the previous studies, MMP9 has been considered to promote the invasion and metastasis of HCC.28 However, whether GIT1 is implicated in regulating the ERK signaling and MMP9 expression in HCC has not been clarified.
In the present study, we identified that GIT1 is an independent prognostic biomarker for predicting both the overall survival and disease-free survival of HCC patients. We found that downregulation of GIT1 facilitated HCC cell apoptosis and repressed HCC cell invasion, migration, and proliferation. Overexpression of GIT1 is associated with phospho (p)-ERK1/2 amplification in HCC tissues. Moreover, downregulation of GIT1 resulted in inactivation of ERK signaling and downregulation of MMP9. Noteworthy, our findings indicate that GIT1 is a candidate oncogene in HCC because it plays a critical role in the initiation and progression of HCC.
Materials and methods
Cell lines and cell culture
Five HCC cell lines (Huh-7, HepG2, SMMC-7721, PLC/PRF5, and MHCC-97H) were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, People’s Republic of China). The immortalized human liver cell line L02 was purchased from the American Type Culture Collection (Manassas, VA, USA). All the cell lines were maintained in complete Dulbecco’s Modified Eagle’s Medium (DMEM) (Invitrogen Life Technologies, Carlsbad, CA, USA) with 10% fetal bovine serum (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) and cultured in a humidified incubator containing 5% CO2 at 37°C.
Clinical samples
A total of 130 pairs of HCC and corresponding normal tumor-adjacent samples (>2.0 cm distance from the margin of the resection) were obtained from the patients who underwent surgical resection between January 2007 and January 2010 in the Eastern Hepatobiliary Surgery Hospital of Second Military Medical University. HCC stages were classified according to the seventh edition tumor-node-metastasis (TNM) classification of the American Joint Committee on Cancer/International Union Against Cancer.29 The clinicopathologic data of these HCC patients are shown in Table 1. All tissues were collected immediately after surgical resection, snap-frozen in liquid nitrogen, and stored at -80°C until use. All recruited HCC patients provided written informed consent before hepatectomy, and all the protocols were approved by the Ethics Committee of the Eastern Hepatobiliary Surgery Hospital of Second Military Medical University according to the 1975 Declaration of Helsinki.
Quantitative reverse transcription-polymerase chain reaction
Total RNA was isolated from cell lines and liver specimens using TRIzol reagent (Invitrogen Life Technologies). RNA samples were reverse transcribed into cDNA with a RevertAid Premium First Strand cDNA Synthesis Kit (Fermentas, Burlington, Canada). Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was done in an ABI 7500 system using the SYBR Premix Ex Taq II (TakaRa, Kyoto, Japan). The primer sequences were: GIT1 sense primer: 5′-CTGAGGATGTCCCGAAAGG-3′; antisense primer: 5′-CAGCACACCCCTGCTGAT-3′; β-actin sense primer: 5′-GGGAAATCGTGCGTGACAT-3′; and antisense primer: 5′-CTGGAAGGTGGACAGCGAG-3′. The experiments were performed in triplicate.
Western immunoblotting
The following primary antibodies were used in the western immunoblotting assays: ERK1/2 (ab17942, Abcam, Cambridge, UK), p-ERK1/2 (ab47339, Abcam), GIT1 (sc-9657, Santa Cruz Biotechnology, Santa Cruz, CA, USA), MMP9 (sc-10737, Santa Cruz Biotechnology), and β-actin (sc-130656, Santa Cruz Biotechnology). The blots were detected with the secondary antibody conjugated with horseradish peroxidase and reactions were visualized using the HyGLO horseradish peroxidase detection kit from Denville (Metuchen, NJ, USA).
Immunohistochemical staining
Immunohistochemical staining was performed on paraformaldehyde-fixed paraffin sections. The primary antibodies GIT1 (sc-9657) and p-ERK1/2 (ab47339) were used in the immunohistochemistry assays. Immunohistochemical staining was performed as previous study reported.6 Semi-quantitative scores were used to assess immunostaining of each HCC sample. Intensity of staining was evaluated as four grades: 0, negative; 1, weak; 2, moderate; 3, strong. And the percentage of positive cells was categorized as the following grades: 0, 0%; 1, 1%–10%; 2, 11%–50%; 3, 51%–80%; and 4 >80%. The staining intensity and average percentage of positive cells were assessed for ten independent high magnification fields. By multiplying the immunostaining intensity and the percentage of positive cells, the final score was obtained (0–12).
RNA interference transfection
The specific small interfering RNA (siRNA) sequences against GIT1 (sc-35477) and control siRNA (sc-37007) were both purchased from Santa Cruz Biotechnology. HCC cells were seeded at a concentration of 2×106 per well in six-well plates and cultured overnight. Next, HCC cells were transfected with 100 nM siRNA using Lipofectamine RNA interference MAX Reagent (Invitrogen Life Technologies) according to the manufacturer’s instructions. Then, HCC cells were used for further experiments at 48 hours after transfection.
Cell proliferation and viability assays
For the tumor cell proliferation assay, HCC cells were seeded into 96-well plates at 5×103 cells per well for 24 hours and determined using a Cell Proliferation ELISA, 5-bromodeoxyuridine (BrdU) (chemiluminescent) (Roche, Indianapolis, IN, USA). Briefly, HCC cells were incubated with BrdU labeling solution for 2 hours, then tumor cells were incubated with peroxidase-conjugated anti-BrdU antibody. The viability of tumor cells was performed using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. HCC cells were seeded in a 96-well plate. The absorbance of the samples was detected at 24, 48, and 72 hours.
Cell apoptotic assays
For the HCC cell apoptosis assay, an Annexin-V-FLUOS Staining Kit (Roche Diagnostics, Mannheim, Germany) was used to determine the level of tumor cell apoptosis, according to the manufacturer’s protocols. The samples were assessed with a BD FACS Canto II Flow Cytometer (Becton Dickinson, Franklin Lakes, NJ, USA). The caspase 3/7 activity assay was determined using an Apo-ONE Homogeneous Caspase-3/7 Assay (Promega, Madison, WI, USA).
Cell invasion and migration assays
Transwell migration assays were performed in 12-well plates with 8 μm BioCoat control inserts (BD Biosciences, Bedford, MA, USA). A concentration of 2×105 GIT1-siRNA or control-siRNA transfected MHCC-97H cells that were suspended in 500 μL serum-free DMEM were added into the upper chamber and DMEM medium with 10% fetal bovine serum was added into the lower chamber. After completion, the HCC cells on the inner layer were removed and the adherent HCC cells on undersurface of the insert were stained with crystal violet. At least five representative images of each well were taken and HCC cell numbers were counted using ImageJ. BioCoat Matrigel invasion chamber (Becton, Dickinson and Company, Bedford, MA) was used for Transwell invasion assay and the following protocols were the same as migration assay.
Statistical analysis
The SPSS 16.0 statistical software package (SPSS Inc., Chicago, IL, USA) was used for statistical analysis. A two-tailed Student’s t-test, a Kaplan–Meier plot, a log-rank test, a chi-square test, or Fisher’s exact test was used to assess statistical significance. Independent prognostic factors were evaluated by the Cox proportional hazards stepwise regression model. Data were presented as the mean ± standard error of the mean. P-values were two-sided, and P<0.05 was considered to be statistically significant.
Results
Upregulation of GIT1 is a frequent event in HCC lines
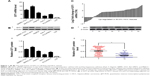
GIT1 expression was first detected in an immortalized nontumorigenic human liver cell line L02 and a panel of HCC cell lines (Huh-7, HepG2, SMMC-7721, PLC/PRF5, and MHCC-97H) by Western blotting and qRT-PCR. We found that the GIT1 mRNA expression was significantly upregulated in all HCC cell lines as compared with that in an immortalized nontumorigenic human liver cell line L02 (P<0.01, respectively, Figure 1A). The results of Western blotting assay also verified these findings (Figure 1B). Our data indicate that overexpression of GIT1 is a frequent event in HCC lines. Moreover, MHCC-97H expressed the highest expression level of GIT1 in all HCC cell lines (Figure 1A and B). Thereby, MHCC-97H cell line was used in further GIT1 knockdown experiments.
Clinical significance of GIT1 expression in HCC samples
To investigate the clinical significance of GIT1 in HCC, we detected the GIT1 expression level in 130 pairs of HCC and matched tumor-adjacent noncancerous samples using qRT-PCR and immunoblotting, and found that the GIT1 expression was evidently higher in the HCC samples than that in the noncancerous samples (P<0.01, respectively, Figure 1C and D). To further determine the clinical role of GIT1 in HCC, we tried to explore the correlations of the GIT1 expression level with patients’ clinicopathological characteristics, including sex, age, hepatitis B surface antigen (HBsAg) status, tumor size, cirrhosis, α-fetoprotein expression, capsule formation, Edmondson–Steiner grade, vascular invasion, tumor number, and TNM stage. The median GIT1 protein expression was used as the cutoff point to divide into low-expressing GIT1 and high-expressing GIT1 groups. In our study, the expression level of GIT1 was significantly correlated with the Edmondson–Steiner grade, tumor number, vascular invasion, and TNM stage (P<0.05, respectively). However, no evident correlation was found between the expression of GIT1 and sex, age, HBsAg status, tumor size, cirrhosis, α-fetoprotein expression, and capsule formation (P>0.05, respectively). Our results are listed in Table 1.
GIT1 expression is an independent prognostic factor for HCC
In this study, the median GIT1 protein expression was used as the cutoff point to divide into low-expressing GIT1 and high-expressing GIT1 groups for patients’ survival. The HCC patients with high GIT1 expression had significantly reduced overall survival and disease-free survival. The 5-year overall survival rate of the high-expressing GIT1 group was 32.3%, which was significantly lower than that of the low-expressing GIT1 group (66.2%, P<0.01) (Figure 2A). The 5-year disease-free survival rate of the high-expressing GIT1 group was 13.8%, which was evidently lower than that of the low-expressing GIT1 group (50.4%, P<0.01) (Figure 2B). Furthermore, univariate analysis revealed that HBsAg status, tumor number, vascular invasion, Edmonson–Steiner classification, TNM stage, and GIT1 expression level were the prognostic factors for the patients with HCC (Table 2). In a multivariate analysis model, the GIT1 expression level was significantly associated with overall survival (hazard ratio 2.301; 95% confidence interval 1.338–3.958; P<0.01) and disease-free survival (hazard ratio 2.106; 95% confidence interval 1.322–3.355; P<0.01) (Table 3). These results indicated that the expression level of GIT1 was an independent prognostic factor of the patients with HCC.
Knockdown of GIT1-promoted HCC cell apoptosis and repressed proliferation invasion and migration
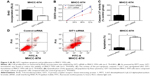
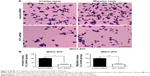
To further explore the effect of GIT1 on HCC cell apoptosis, proliferation, invasion, and migration, we downregulated the GIT1 expression using GIT1-siRNA in MHCC-97H cells. As compared with the control group, knockdown of GIT1 expression was found to inhibit HCC cells proliferation and viability markedly by BrdU and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assays (P<0.01, respectively, Figure 3A and B). Furthermore, our study revealed that the activity of caspase 3/7 in MHCC-97H cells evidently increased after downregulation of GIT1 expression (P<0.01, Figure 3C). Flow cytometry assay showed that the percentage of apoptosis cells in GIT1-siRNA group increased (P<0.01, respectively, Figure 3D). In this study, we also found that downregulation of GIT1 expression markedly reduced the number of migrated and invaded HCC cells (P<0.01, respectively, Figure 4A and B).
Correlation between GIT1 and p-ERK1/2 protein expression
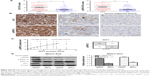
To explore whether GIT1 overexpression is associated with ERK activation, we detected the expression level of GIT1 and p-ERK1/2 using immunohistochemical staining in HCC tissues and matched tumor-adjacent liver tissues. In our study, both GIT1 and p-ERK1/2 levels were found to be evidently higher in HCC tissues as compared with those in corresponding adjacent no tumorous liver tissues (Figure 5A and B). And there was indeed a significant positive correlation between the expression level of GIT1 and p-ERK1/2 (r=0.592, P<0.01, Spearman’s correlation test) (Figure 5C).
Knockdown of GIT1 was associated with inactivation of ERK signaling and downregulation of MMP9 in HCC cells
To investigate the underlying mechanism by which GIT1 regulates HCC cell apoptosis, proliferation, invasion, and migration, we detected the effect of GIT1 on ERK signaling. In this study, downregulation of GIT1 expression markedly attenuated the phosphorylation of ERK1/2 in MHCC-97H cells (P<0.01, Figure 5D). Moreover, to better understand the downstream molecules implicated in GIT1-induced ERK signaling activation, we detected whether GIT1 affected MMP9 expression in MHCC-97H cells. As expected, we found that downregulation of GIT1 expression evidently decreased the MMP9 expression level in MHCC-97H cells (P<0.01, Figure 5D).
To better understand the underlying mechanism of GIT1 in HCC, we further determined whether ERK phosphorylation status affected GIT1/ERK/MMP9 signaling activation in HCC cells. In this study, we found that knockdown of GIT1 had no effect on MMP9 expression after inhibiting ERK phosphorylation using ERK inhibitor SCH772984 (sc-473205, Santa Cruz Biotechnology) in MHCC-97H cells (Figure 5D). Our data revealed that the GIT1-induced MMP9 overexpression was strongly dependent on the ERK phosphorylation and GIT1-regulated MMP9 expression through ERK signaling in HCC cells.
Discussion
Due to the high frequency of cancer recurrence, the prognosis of HCC patients remains poor.4 Recent studies have revealed many aberrantly expressed genes contributing to the progression of HCC, but the molecules that help to predict tumor recurrence and serve as potential targets remain very limited.30–32 Thus, it is urgent to identify novel biomarkers that are useful to improve the treatment efficacy of HCC. GIT1, a scaffold protein, is considered to be involved in regulating several aspects of cancer progression.11,19 Several studies have indicated that GIT1 overexpression is commonly observed in a wide variety of human tumors.14,21 Additionally, it has been revealed that GIT1 can facilitate tumor progression via activating ERK signaling in human cancers.22
The role of GIT1 has been investigated in many cancer types.14,17,20 However, the GIT1 expression and its related mechanisms in HCC remained unclear. In our study, we initially detected the expression level of GIT1 in a nontumorigenic hepatocyte cell line L02 and five hepatoma cell line (Huh-7, HepG2, SMMC-7721, PLC/PRF5, and MHCC-97H). We found that GIT1 expression level in hepatoma cell lines was markedly higher than that in nontumorigenic hepatocyte cell line L02. We further detected GIT1 expression in 130 HCC samples using qRT-PCR, Western blotting, and immunohistochemical staining, and our results revealed that the GIT1 expression level was obviously higher in HCC samples compared with matched normal tumor adjacent liver samples. Furthermore, high GIT1 expression level was evidently correlated with several clinicopathological parameters including Edmondson–Steiner grade, tumor number, vascular invasion, and TNM stage. Notably, our results also demonstrate that high-expression level of GIT1 was obviously correlated with shorter overall survival and disease-free survival, which is an assumed molecular marker for negative prognosis in patients with HCC. Multivariate Cox repression analysis further demonstrated that GIT1 is an independent factor for patients with HCC. Taken together, these results confirmed that the expression level of GIT1 may facilitate tumor progression and is an important factor for prognosis determination in patients with HCC.
In this study, we identified the role of GIT1 in HCC cells. We found that downregulation of GIT1 expression obviously promoted tumor cell apoptosis and repressed proliferation invasion and migration in MHCC-97H cells. The in vitro experiments presented in our study demonstrate that GIT1 is a critical regulatory factor in HCC cell apoptosis, proliferation, invasion, and migration. In our study, we also detected the expression level of GIT1 and p-ERK1/2 using immunohistochemical staining in HCC tissues and matched tumor-adjacent liver tissues. As expected, there was indeed an evident positive correlation between the expression of GIT1 and p-ERK1/2 in HCC tissues. Our data indicate that the overexpression of GIT1 can cause ERK signaling activation in HCC.
The ERK signaling is an important signal transduction pathway that affects many cellular processes, such as cell apoptosis, proliferation, invasion, and migration.24 Mounting evidence has revealed that the activation of ERK signaling is believed to contribute to human carcinogenesis.23,31 Importantly, ERK activation was recently found to be critical for the overexpression of MMPs in human cancer.26 In this study, we detected the GIT1 expression and its correlation with ERK activation in HCC cells. Downregulation of GIT1 expression evidently decreased the expression of p-ERK1/2. These data suggest that GIT1 regulates HCC cell apoptosis, proliferation, invasion, and migration via stimulating ERK signaling activity.
The overexpression of MMPs has been related to tumor cell migration and invasion.28,29 MMPs are largely involved in promoting tumorigenesis in multiple cancer types including HCC.26,28 As a member of MMPs, MMP9 is a critical factor in regulating the migration and invasion of HCC.31 Considering the fact that an important biological function of ERK signaling is to regulate the expression of MMP9, we identified whether GIT1 affected ERK/MMP9 signaling in HCC cells.24,27 We found that downregulation of GIT1 expression obviously decreased the expression of p-ERK1/2 and MMP9. Furthermore, GIT1-induced MMP9 overexpression was strongly dependent on the ERK phosphorylation and GIT1-regulated MMP9 expression through ERK signaling in HCC cells. Our data confirmed that GIT1 mediates the activation of ERK and upregulation of MMP9 and GIT1-induced ERK/MMP9 signaling activation may be responsible for the progression of HCC.
Conclusion
We demonstrated that GIT1 expression level is upregulated in HCC tissues as compared with noncancerous liver tissues and high-expression level of GIT1 is correlated with clinicopathological characteristics of poor prognosis in patients with HCC. Furthermore, we found that GIT1 expression level is an independent factor for predicting the overall survival and disease-free survival of HCC patients. Notably, GIT1 can repress cell apoptosis and promote cell proliferation, invasion, and migration via activating ERK/MMP9 signaling in HCC. This study provides a better understanding on both the clinical role and molecular biological mechanism of GIT1 in HCC. Our current work demonstrated that GIT1 has an oncogenic role in hepatocarcinogenesis and promotes HCC progression via activating ERK/MMP9 signaling pathway.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (81372674). The authors thank all the patients who participated in this study.
Disclosure
The authors report no conflicts of interest in this work.
References
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. | ||
Lafaro KJ, Demirjian AN, Pawlik TM. Epidemiology of hepatocellular carcinoma. Surg Oncol Clin N Am. 2015;24(1):1–17. | ||
Bosetti C, Levi F, Boffetta P, Lucchini F, Negri E, La Vecchia C. Trends in mortality from hepatocellular carcinoma in Europe, 1980–2004. Hepatology. 2008;48(1):137–145. | ||
Bosetti C, Turati F, La Vecchia C. Hepatocellular carcinoma epidemiology. Best Pract Res Clin Gastroenterol. 2014;28(5):753–770. | ||
Kee KM, Hung CH, Wang JH, Lu SN. Serial changes of clinical parameters in a patient with advanced hepatocellular carcinoma with portal vein thrombosis achieving complete response after treatment with sorafenib. Onco Targets Ther. 2014;7:829–834. | ||
Liu Q, Tu K, Zhang H, Zheng X, Yao Y, Liu Q. TPX2 as a novel prognostic biomarker for hepatocellular carcinoma. Hepatol Res. 2015;45(8):906–918. | ||
Park JW, Chen M, Colombo M, et al. Global patterns of hepatocellular carcinoma management from diagnosis to death: the BRIDGE Study. Liver Int. 2015;35(9):2155–2166. | ||
Lu M, Li J, Luo Z, et al. Roles of dopamine receptors and their antagonist thioridazine in hepatoma metastasis. Onco Targets Ther. 2015;8:1543–1552. | ||
Du R, Wu S, Lv X, Fang H, Wu S, Kang J. Overexpression of brachyury contributes to tumor metastasis by inducing epithelial-mesenchymal transition in hepatocellular carcinoma. J Exp Clin Cancer Res. 2014;33:105. | ||
Penela P, Nogues L, Mayor F Jr. Role of G protein-coupled receptor kinases in cell migration. Curr Opin Cell Biol. 2014;27:10–17. | ||
Majumder S, Sowden MP, Gerber SA, et al. G-protein-coupled receptor-2-interacting protein-1 is required for endothelial cell directional migration and tumor angiogenesis via cortactin-dependent lamellipodia formation. Arterioscler Thromb Vasc Biol. 2014;34(2):419–426. | ||
Yoo SM, Antonyak MA, Cerione RA. The adaptor protein and Arf GTPase-activating protein Cat-1/Git-1 is required for cellular transformation. J Biol Chem. 2012;287(37):31462–31470. | ||
Dent LG, Poon CL, Zhang X, et al. The GTPase regulatory proteins Pix and Git control tissue growth via the Hippo pathway. Curr Biol. 2015;25(1):124–130. | ||
Huang WC, Chan SH, Jang TH, et al. miRNA-491-5p and GIT1 serve as modulators and biomarkers for oral squamous cell carcinoma invasion and metastasis. Cancer Res. 2014;74(3):751–764. | ||
Schmalzigaug R, Garron ML, Roseman JT, et al. GIT1 utilizes a focal adhesion targeting-homology domain to bind paxillin. Cell Signal. 2007;19(8):1733–1744. | ||
Zhao ZS, Manser E, Loo TH, Lim L. Coupling of PAK-interacting exchange factor PIX to GIT1 promotes focal complex disassembly. Mol Cell Biol. 2000;20(17):6354–6363. | ||
Chan SH, Huang WC, Chang JW, et al. MicroRNA-149 targets GIT1 to suppress integrin signaling and breast cancer metastasis. Oncogene. 2014;33(36):4496–4507. | ||
Li J, Wang Q, Wen R, et al. MiR-138 inhibits cell proliferation and reverses epithelial-mesenchymal transition in non-small cell lung cancer cells by targeting GIT1 and SEMA4C. J Cell Mol Med. 2015. | ||
Huang C, Sheng Y, Jia J, Chen L. Identification of melanoma biomarkers based on network modules by integrating the human signaling network with microarrays. J Cancer Res Ther. 2014;10 Suppl:C114–C124. | ||
Zhang N, Cai W, Yin G, Nagel DJ, Berk BC. GIT1 is a novel MEK1-ERK1/2 scaffold that localizes to focal adhesions. Cell Biol Int. 2010;34(1):41–47. | ||
Peng H, Dara L, Li TW, et al. MAT2B-GIT1 interplay activates MEK1/ERK 1 and 2 to induce growth in human liver and colon cancer. Hepatology. 2013;57(6):2299–2313. | ||
Peng H, Li TW, Yang H, Moyer MP, Mato JM, Lu SC. Methionine adenosyltransferase 2B-GIT1 complex serves as a scaffold to regulate Ras/Raf/MEK1/2 activity in human liver and colon cancer cells. Am J Pathol. 2015;185(4):1135–1144. | ||
Zheng F, Wu J, Zhao S, et al. Baicalein increases the expression and reciprocal interplay of RUNX3 and FOXO3a through crosstalk of AMPKalpha and MEK/ERK1/2 signaling pathways in human non-small cell lung cancer cells. J Exp Clin Cancer Res. 2015;34:41. | ||
Wang ZL, Fan ZQ, Jiang HD, Qu JM. Selective Cox-2 inhibitor celecoxib induces epithelial-mesenchymal transition in human lung cancer cells via activating MEK-ERK signaling. Carcinogenesis. 2013;34(3):638–646. | ||
Xie B, Lin W, Ye J, et al. DDR2 facilitates hepatocellular carcinoma invasion and metastasis via activating ERK signaling and stabilizing SNAIL1. J Exp Clin Cancer Res. 2015;34(1):101. | ||
Kim HC, Kim YS, Oh HW, et al. Collagen triple helix repeat containing 1 (CTHRC1) acts via ERK-dependent induction of MMP9 to promote invasion of colorectal cancer cells. Oncotarget. 2014;5(2):519–529. | ||
Lin F, Chengyao X, Qingchang L, Qianze D, Enhua W, Yan W. CRKL promotes lung cancer cell invasion through ERK-MMP9 pathway. Mol Carcinog. 2015;54 Suppl 1:E35–E44. | ||
Liu Q, Yang P, Tu K, et al. TPX2 knockdown suppressed hepatocellular carcinoma cell invasion via inactivating AKT signaling and inhibiting MMP2 and MMP9 expression. Chin J Cancer Res. 2014;26(4):410–417. | ||
Wu Z, Jiang P, Zulqarnain H, Gao H, Zhang W. Relationship between single-nucleotide polymorphism of matrix metalloproteinase-2 gene and colorectal cancer and gastric cancer susceptibility: a meta-analysis. Onco Targets Ther. 2015;8:861–869. | ||
Muntane J, De la Rosa AJ, Docobo F, Garcia-Carbonero R, Padillo FJ. Targeting tyrosine kinase receptors in hepatocellular carcinoma. Curr Cancer Drug Targets. 2013;13(3):300–312. | ||
Whittaker S, Marais R, Zhu AX. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene. 2010;29(36):4989–5005. | ||
Xie B, Zen Q, Wang X, et al. ACK1 promotes hepatocellular carcinoma progression via downregulating WWOX and activating AKT signaling. Int J Oncol. 2015;46(5):2057–2066. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.