Back to Journals » Infection and Drug Resistance » Volume 13
Genotypic to Phenotypic Resistance Discrepancies Identified Involving β-Lactamase Genes, blaKPC, blaIMP, blaNDM-1, and blaVIM in Uropathogenic Klebsiella pneumoniae
Authors Urmi UL , Nahar S, Rana M , Sultana F , Jahan N , Hossain B , Alam MS, Mosaddek ASM , McKimm J , Rahman NAA , Islam S , Haque M
Received 12 May 2020
Accepted for publication 16 July 2020
Published 18 August 2020 Volume 2020:13 Pages 2863—2875
DOI https://doi.org/10.2147/IDR.S262493
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Umme Laila Urmi,1 Shamsun Nahar,1 Masud Rana,1 Fahmida Sultana,1 Nusrat Jahan,1 Billal Hossain,1 Mohammed Shah Alam,2 Abu Syed Md Mosaddek,3 Judy McKimm,4 Nor Azlina A Rahman,5 Salequl Islam,1 Mainul Haque6
1Department of Microbiology, Jahangirnagar University, Dhaka 1342, Bangladesh; 2Department of Microbiology, Gono Bishwabidyalay, Savar, Dhaka 1342, Bangladesh; 3Department of Pharmacology, Uttara Adhunik Medical College, Uttara, Dhaka, Bangladesh; 4Medical Education, Strategic Educational Development, Leadership for the Health Professions, Swansea University Medical School, Swansea University, Swansea, Wales SA2 8PP, UK; 5Department of Physical Rehabilitation Sciences, Kulliyyah of Allied Health Sciences, International Islamic University Malaysia, Kuantan 25200, Malaysia; 6Faculty of Medicine and Defence Health, Universiti Pertahanan Nasional Malaysia (National Defence University of Malaysia), Kuala Lumpur 57000, Malaysia
Correspondence: Salequl Islam
Department of Microbiology, Jahangirnagar University, Savar, Dhaka 1342, Bangladesh
Tel +880-1715029136
Fax +880-2-7791052
. Email [email protected]
Mainul Haque
Faculty of Medicine and Defence Health, Universiti Pertahanan Nasional Malaysia (National Defence University of Malaysia), Kem Sungai Besi, Kuala Lumpur 57000, Malaysia
Tel +60 10 926 5543
Email [email protected]
Introduction: Klebsiella pneumoniae carbapenemase (KPC) belongs to the Group-A β-lactamases that incorporate serine at their active site and hydrolyze various penicillins, cephalosporins, and carbapenems. Metallo-beta-lactamases (MBLs) are group-B enzymes that contain one or two essential zinc ions in the active sites and hydrolyze almost all clinically available β-lactam antibiotics. Klebsiella pneumoniae remains the pathogen with the most antimicrobial resistance to KPC and MBLs.
Methods: This research investigated the blaKPC, and MBL genes, namely, blaIMP, blaVIM, and blaNDM-1 and their phenotypic resistance to K. pneumoniae isolated from urinary tract infections (UTI) in Bangladesh. Isolated UTI K. pneumoniae were identified by API-20E and 16s rDNA gene analysis. Their phenotypic antimicrobial resistance was examined by the Kirby-Bauer disc diffusion method, followed by minimal inhibitory concentration (MIC) determination. blaKPC, blaIMP, blaNDM-1, and blaVIM genes were evaluated by polymerase chain reactions (PCR) and confirmed by sequencing.
Results: Fifty-eight K. pneumoniae were identified from 142 acute UTI cases. Their phenotypic resistance to amoxycillin-clavulanic acid, cephalexin, cefuroxime, ceftriaxone, and imipenem were 98.3%, 100%, 96.5%, 91.4%, 75.1%, respectively. Over half (31/58) of the isolates contained either blaKPC or one of the MBL genes. Individual prevalence of blaKPC, blaIMP, blaNDM-1, and blaVIM were 15.5% (9), 10.3% (6), 22.4% (13), and 19% (11), respectively. Of these, eight isolates (25.8%, 8/31) were found to have two genes in four different combinations. The co-existence of the ESBL genes generated more resistance than each one individually. Some isolates appeared phenotypically susceptible to imipenem in the presence of blaKPC, blaIMP, blaVIM, and blaNDM-1 genes, singly or in combination.
Conclusion: The discrepancy of genotype and phenotype resistance has significant consequences for clinical bacteriology, precision in diagnosis, the prudent selection of antimicrobials, and rational prescribing. Heterogeneous phenotypes of antimicrobial susceptibility testing should be taken seriously to avoid inappropriate diagnostic and therapeutic decisions.
Keywords: Klebsiella pneumoniae, blaKPC, blaIMP, blaNDM-1, blaVIM, co-resistance, heteroresistance, urinary tract infections, Bangladesh
Introduction
Currently, β-lactams are the most extensively used antibiotics, which include natural and synthetic penicillins and their derivatives such as cephalosporins, cephamycins, monobactams, and carbapenems.1 Bacterial β-lactamase enzymes that hydrolyze the β-lactam ring inactivate the drugs, thereby conferring resistance. Biochemically β-lactamase enzymes are classified into two broad groups. The first group of enzymes incorporate serine at their active site and hydrolyze the amide bond in the lactam ring of antibiotics to render it inactive.2 The second group is metallo-β-lactamases (MBLs) that contain one or two essential zinc ions in the active sites and facilitate a hydrolytic reaction to target drugs.3 The widely known Ambler classification of β-lactamases divides these enzymes into four molecular classes (A, B, C, and D) based upon the amino acid sequences and molecular homology between active site amino acid motifs.4 Klebsiella pneumoniae carbapenemase (KPC) belongs to the Group-A β-lactamases, which hydrolyze penicillins, cephalosporins, and carbapenems,5,6 and is inhibited partially by class A inhibitors such as clavulanate or tazobactam.7 Group C comprises cephalosporinases, including the CMY-family AmpC,8,9 and Group D are oxacillinase, consisting of OXA-48, OXA-23, and similar enzymes.10,11 Unlike the serine-active proteins, MBLs exhibit wide-spectrum hydrolysis in all beta-lactams except aztreonam, also clavulanate or tazobactam cannot repress MBLs.12 Among nine types of reported MBLs, blaIMP, blaNDM, and blaVIM are the most prevalent genes worldwide, including in Africa, Asia, Europe, and the Americas.13 blaIMP-1 emerged and spread during the early 1990s in Japan then was found in other countries.14 The blaVIM (Verona Integron-encoded Metallo β-lactamase) gene was first found in Europe and then emerged in other countries. However, blaNDM-producing bacteria were first isolated from a Swedish resident who contracted a urinary tract infection caused by carbapenem-resistant K. pneumoniae while he was in New Delhi in late 2007.15 Subsequently, the blaNDM gene emerged in Pakistan, the Indian subcontinent and the United Kingdom.16,17 Multiple resistance genes in the same bacterial isolate accumulate some compound phenotype known as co-resistance, which was reported among ESBLs with an increased opportunity of transmission.18
Gram-negative bacteria, particularly carbapenem-resistant Enterobacteriaceae (CRE), currently pose a severe global human health threat.19,20 As such, the World Health Organization (WHO) identified and prioritized CRE as one of the most critical antibiotic-resistant bacteria (WHO priority list 2017). Antibiotic resistance in CRE is, therefore, a vital area for clinical and public health research.21 Most Enterobacteriaceae are healthy flora; however, some become pathogens, causing a diversified severe infection, including systemic bacteremia, community-acquired infections, healthcare-associated infections (HAIs), and both complicated and uncomplicated urinary tract infections (UTIs).22,23 Klebsiella pneumoniae remains the leading pathogen producing carbapenemase24 and MBL enzymes17,25 and has been reported to show co-resistance through containing multiple ESBL genes simultaneously.26 Some antibiotic resistance genes (ARGs) accumulate multiple random mutations in their gene sequences over time and become dysfunctional resistance genes (resistance pseudogenes) that fail to exhibit expected resistance phenotypes.27 The pseudo-ARGs are often considered to comprise nonfunctional junk DNA that remains stable in the bacterial genome or plasmid.28 They become attenuated or impaired in their ability to produce functional β-lactamases, leading to phenotype-genotype discrepancies.
The present study investigated the presence of serine β-lactamase gene, KPC, and MBL genes, such as blaIMP, blaVIM, and blaNDM-1 in Klebsiella pneumoniae isolates from UTI patients in Dhaka, Bangladesh. The study also analyzed the AMR genotypes and phenotypes of the UTI isolates among the four ESBL genes and different β-lactam antibiotics.
Materials and Methods
Study Design and Specimen Collection
A cross-sectional study was conducted between April 2017 and March 2018 among patients showing clinical signs of a UTI who attended outpatient Departments at Gonosastha Medical College Hospital, and Enam Medical College Hospital in Savar, Dhaka, Bangladesh. Only patients with no history of antibiotic treatment in the previous 15 days were invited to participate in the study. Patients with a known record of immunocompromised diseases, including cancer, organ transplant, HIV/AIDS, and renal disorders were also excluded from the study. Informed written consent was obtained from each study participant prior to collecting their demographic data and urine specimens. For participants who were minors, consent was obtained from their parents or legal guardians. The patients’ main presenting clinical signs and symptoms were recorded using a structured questionnaire. All 142 patients met the study criteria for microbiological investigation. The standard clean-catch midstream urine sampling procedure was explained to the participants, including the importance of avoiding contamination with commensal bacteria,29 then the samples were collected and immediately transferred to the laboratory for analysis.
Bacterial Isolation and Identification
Urine samples collected in sterile glass tubes were inoculated on differential culture media, cysteine-, lactose-, and electrolyte-deficient (CLED) agar (Lyophilchem, Italy) and MacConkey agar. The specimens were plated within two hours after collection to avoid false-positive results. Using CLED agar medium inhibits the swarming growth of Proteus species and other gram-negative bacilli in favor of gram-positive cocci if present in the specimen. Urine cultures were incubated overnight at 35ºC–37ºC in ambient air. Quantitative urine cultures were carried out, and colony counts of 102 or 103 CFU/mL were considered to define a probable UTI infection.30 Colony counts of less than 102 CFU/mL were assumed as potentially contaminated. The isolates were sub-cultured on nutrient agar (Oxoid, UK), and their identification was initially performed by Gram’s staining and biochemical tests, including the oxidase test, Kligler iron agar (KIA), Indole test, citrate utilization, urease test, and motility test. Identification was confirmed by a rapid biochemical-test kit (API 20E, BioMe´rieux, Durham, NC) consisting of a set of chromogenic panels, carbohydrate batteries, and enzymatic substrates.31 The bacterial identity was validated further by the amplification and sequencing of the 16S rDNA gene.32 The isolates were preserved in 30% glycerol at - 20ºC in Trypticase Soy Broth (TSB) until further analysis. This study included only the exclusive Klebsiella pneumoniae isolates from UTI patients of all age groups and both sexes. Samples infected with co-pathogens were excluded from the analyses.
Antimicrobial Susceptibility Testing
The phenotypic antimicrobial susceptibilities of the isolates were tested by the disc diffusion method (Kirby–Bauer) on Mueller–Hinton agar (Oxoid, Basingstoke, UK) plates according to the Clinical and Laboratory Standards Institute (CLSI) guidelines.33 Briefly, a 4-hour bacterial suspension in Mueller–Hinton broth was adjusted to a density of McFarland 0.5 equivalent and then evenly streaked on MHA plates to ensure consistent growth. Antibiotic discs were placed on the bacterial lawn and incubated at 37ºC overnight to determine the sensitivity pattern. Sensitive bacteria developed a clear zone around each disc, and zone diameter was measured and evaluated. Escherichia coli ATCC25922 was used as the susceptible-control reference strain for disc diffusion testing. Antibiotic discs were procured from Oxoid limited (Basingstoke, UK). The β-lactam antibiotics tested were as follows: Amoxycillin+Clavulinic acid (30 µg), Cephalexin (30 µg), Cefuroxime Sodium (30 µg), Ceftriaxone (30 µg), Cefepime (30 µg), and Imipenem (10 µg). Outside the β-lactam group, two commonly prescribed antibiotics for UTI, namely, trimethoprim/sulfamethoxazole (25 µg) and ciprofloxacin (5 µg) were also included for susceptibility assessment.
Minimum Inhibitory Concentration (MIC) Determination
Resistant isolates from the Kirby-Bauer test were further analysed to determine the lowest concentration of antibiotics required to inhibit their visible growth (minimal inhibitory concentration, MIC). The agar microdilution method was employed primarily to check the MIC.13,34 Etest was carried out on imipenem only in parallel to validate the former MIC results.35 In the procedure, commercial Etest strips carrying concentration gradient (from 0.016 to 256 μg/mL) of imipenem was examined (BioMe´rieux, Durham, NC) according to the manufacturer’s instructions. MH agar plates were inoculated with standardized (0.5 McFarland Standard) saline suspension of each isolate prepared from overnight cultures on MH agar plates. Etest strips were carefully layered on each inoculated plate, and plates were incubated at 35°C for 18 hours. Results were interpreted for K. pneumoniae, according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST, Version 7.1 2017). The clinical breakpoints for imipenem resistance were considered when the MIC value was > 8 μg/mL.36
PCR of blaKPC, blaIMP, blaNDM-1, and blaVIM Genes
All the isolates were subjected to a polymerase chain reaction (PCR) test for genotypic confirmation of serine β-lactamase gene, KPC, and MBL genes, including blaIMP, blaVIM, and blaNDM-1. The total DNA of all the bacterial isolates was extracted by the boiling method from bacteria grown on nutrient agar media.15 Specific primer sets for the respective genes were selected based on previous literature37,38 and synthesized from Integrated DNA Technology (IDT, Singapore). For each PCR reaction, prepared bacterial DNA 2.0 µL was added to a 12 µL 2X PCR pre-mixture (GeneON, Germany) and five pmol of each primer (1 µL), and the remaining deionized water to make a final volume of 24 µL. Reactions underwent an initial denaturation at 95°C for 10 min followed by 32 cycles of amplification (Applied Biosystems 2720 Thermal Cycler, Singapore), consisting of denaturation 30s at 94 °C, annealing 30s at 52–56 °C depending on primer sets, extension 1 min at 72°C, and a final 7 min extension at 72°C. Amplicons were visualized under UV light after electrophoresis through 1.2% agarose gel at 100 volts for 30 minutes, followed by staining with ethidium bromide. The standard molecular weight marker was run in parallel to measure specific amplicon sizes (GeneRuler, ThermoFisher Scientific, MA).
Statistical Analysis
Data were verified, entered, and subsequently analyzed using IBM SPSS statistics data editor (version 21). Missing data were omitted from the bivariate analysis. Descriptive and inferential statistical procedures were used to describe the UTI Klebsiella pneumoniae and their carriage of ESBL genes and phenotypic attributes. Pearson’s chi-square test was used to test any association between categorical data, and Yate’s correction for continuity was applied where required. The correlation coefficient was calculated to check the strength of the association. A two-tailed p-value was calculated to measure statistical significance.
Ethics Statement
This study was approved by the Ethics and Research Review Committee of the Jahangirnagar University (JU) Faculty of Biological Sciences [No. BBEC, JU/M 2017 3(4) dated 15.03.2017]. This research was carried out from an undertaken capstone project of the University. However, JU did not have a full-fledged hospital, from where required biospecimens could be obtained. Therefore, we made research collaborations with nearby private hospitals, namely, Gonosastha Medical College Hospital and Enam Medical College Hospital. Both the private hospitals had accepted the Ethical permission that was taken from JU. Therefore, we completed this work under a common ethical permission. All the study protocols complied with the Declaration of Helsinki for recruiting human subjects for medical research. Written informed consent was obtained from adult study patients for collecting the urine samples. Separate written informed consent was taken from parents or legal guardians for patients under 18 years of ages. A respective sample identification code was assigned to each sample collected. The personal identities and other information on the participants were strictly anonymised to protect their privacy.
Results
Study Participants and Klebsiella pneumoniae Prevalence
During the one year study period, urine samples of 142 patients attending two hospitals in Dhaka city with acute urinary tract infection (UTI) were examined, and 58 (40.8%) Klebsiella pneumoniae isolates were identified. All the urine samples appeared culture-positive with at least one UTI pathogen. Most of the participants were consistently symptomatic. Most of them reported a continuous urge to urinate, a burning sensation during urination, and an incomplete or empty feeling in their bladder. The mean age (±Standard Deviation) of the study patients with Klebsiella pneumoniae infection was 35.7±15.6 years. The age range was from 4 to 76 years. Further subclassification showed that the 20 to 39 years age-group accounted for 44.8% of infections. Incidence was higher among females, 72.4% (n=42) compared to 27.6% (n=16) in males. However, further analysis shows no significant association between K. pneumoniae growth from the urine cultures with age-group [χ2(2, N=142) =1.536, p=0.464] and sex [χ2(1, N=142) =0.714, p=0.398]. The details of the comparison are shown in Figure 1.
Phenotypic Antimicrobial Susceptibility and Minimum Inhibitory Concentration
In vitro antimicrobial potency of the most frequently prescribed orally administered β-lactam antibiotics was tested based on the disc diffusion method against the 58 UTI Klebsiella pneumoniae. Semi-synthetic penicillin, amoxycillin-clavulanic acid showed very weak potency against the test pathogens, where 98.3% of isolates were resistant. First-generation cephalosporins such as cephalexin were among the least potent antimicrobial, with 100% resistance. The second-generation cephalosporin, cefuroxime sodium, was also ineffective, with 96.6% of the pathogens evaluated being resistant. The extended-spectrum third generation and the fourth generation cephalosporins, such as ceftriaxone and cefepime, showed a negligible functional spectrum of activities against only 8.6% of test pathogens: the remaining 91.4% were resistant. In contrast, a carbapenem antibiotic, imipenem, was moderately active against the uropathogens, with 25.9% (15/58) of isolates appearing susceptible. Both ciprofloxacin and trimethoprim/sulfamethoxazole showed moderate activity with susceptibility outputs of 38% (22/58) and 19% (11/58), respectively. The MIC of imipenem for resistance varied from >8 µg/mL to >256 µg/mL. Thus, the MIC results correlated with the disc-diffusion results. The susceptible control, ATCC strain Bacillus cereus, and imipenem susceptible (by disc diffusion) isolates exhibited a MIC <2 µg/mL. MIC assessment by commercial Etest strips also matched the agar diffusion test findings, therefore the tests validated each other.
Genotypic-Phenotypic Agreement of blaKPC, blaIMP, blaNDM-1, and blaVIM
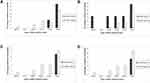
Serine β-lactamase gene blaKPC was detected in 9 (15.5%) UTI K. pneumoniae by PCR. The MBL genes, blaIMP, blaNDM, and blaVIM, were detected in 6 (10.3%), 13 (22.4%), and 11 (19%) isolates, respectively. None of the isolates containing the blaKPC gene was susceptible to Amoxycillin-clavulanic acid, cephalexin, cefuroxime sodium, and ceftriaxone, one isolate (11.1%) showed susceptible to cefepime and three isolates (33.3%) to imipenem. About 75% and 90% of the isolates without harboring the blaKPC gene exhibited resistance to imipenem and ceftriaxone, respectively. The results were similar to other β-lactam antibiotics; however, the degree of resistance was somewhat lower among the KPC-naïve bacteria. Cephalexin (30µg), a first-generation cephalosporin, was found to be ineffective on isolates with or without carrying the blaKPC gene (Figure 2A). Among the blaIMP-positive isolates (n=6), 16.7% sensitivity was observed to Amoxycillin-clavulanic acid, cefuroxime, ceftriaxone, and cefepime. Furthermore, imipenem was found susceptible to 33.3% of IMP-positive and 25% of IMP-negative isolates. Therefore, the presence or absence of the blaIMP gene did not greatly affect phenotypic susceptibility to imipenem. Moreover, some lower susceptibility was observed among IMP-naive pathogens to antibiotics of the tested β-lactam panel (Figure 2B). Among the blaVIM-positive isolates (n=11), none was susceptible to either of the tested Amoxyclav and cephalosporins. blaVIM-negative isolates showed a comparative higher sensitivity, and imipenem remained the most active drug, showing potency to 18.8% of VIM-positive and 27.7% of VIM-negative isolates (Pearson chi-square, p=0.71). In all cases, cephalexin was 100% ineffective on all isolates regardless of the presence of the blaVIM gene (Figure 2C). Among 13 New Delhi Metallo β-lactamase-containing isolates, none was susceptible to Amoxyclav or either of cephalosporins examined, while the blaNDM-negative isolates showed some higher degree of susceptibility. Imipenem remained almost equally sensitive, 23% to NDM-positive, and 27% to NDM-negative isolates (Pearson chi-square, p=1.0), respectively. Consistently, cephalexin remained ineffective regardless of whether the isolates carried the blaNDM-1 gene or not (Figure 2D). The overall presence of the blaKPC gene or MBL genes influenced reduced susceptibility among the UTI K pneumoniae isolates. However, there was no statistically significant association between the antimicrobial susceptibility and the presence of the ESBL genes. The relatively small sample size in this study may have affected the statistical association observed. The difference between phenotypic susceptibility in the presence of the blaKPC and MBL genes was noticeable.
Association Between Phenotypic Antimicrobial Susceptibility with Age and Sex
Fisher’s Exact test was used to assess the association between the phenotypic antimicrobial susceptibility with age and sex because the expected frequency assumption was not sufficient to use the Chi-square test for the majority of antimicrobials. Age and antimicrobial susceptibility were reclassified into two groups, and sensitive and intermediate antimicrobial susceptibility were combined into one group to be compared with the samples resistant to antimicrobials. It was found that none of the phenotypic antimicrobial susceptibility tested on the urine samples with the growth of K. pneumonia showed any significant relationship with either age or sex, with p-values ranging from 0.053 to 1.000. Details of the test are shown in Table 1, except for cephalexin, where the analysis could not be performed because all samples were resistant towards it.
 |
Table 1 Association Between Phenotypic Antimicrobial Susceptibility of Urine K. Pneumoniae with Age and Sex (Fisher’s Exact Test, n=58) |
Association Between Genotypic blaKPC, blaIMP, blaNDM-1, and blaVIM with Age and Sex
As in 2.4, Fisher’s Exact test was used to assess the association between genotypic blaKPC, blaIMP, blaNDM-1, and blaVIM with age and sex except for blaVIM where the Chi-square test was used to compare between different age groups. Similarly, no significant results were found, with p-values ranging between 0.097 and 1.000, as shown in Table 2.
 |
Table 2 Association Between Genotypic blaKPC, blaIMP, blaNDM-1, and blaVIM of Urine K. Pneumoniae with Age and Sex (Fisher’s Exact Test, n=58) |
Co-Resistance Phenotypes of blaKPC, blaIMP, blaNDM-1, and blaVIM
A total of 31 isolates among 58 K. pneumoniae were found to carry at least one of the four ESBL genes. Most of the isolates carried a single gene, although some carried multiple genes simultaneously. Co-existing ESBL genes were found in four combinations: blaVIM +blaKPC, blaVIM +blaIMP, blaVIM +blaNDM-1, and blaNDM-1+blaIMP. bla-VIM had the highest combination of co-existence with KPC, IMP, and NDM. From 11 isolates carrying blaVIM-gene, four were mutually exclusive; five had a concurrence with NDM, one with KPC and one with IMP. Another co-existence was found in one isolate between IMP and NDM (Figure 3). Thus, there were eight isolates (25.8%, 8/31) containing any two of the ESBL genes. The remaining 23 (74.2%) isolates hosted ESBL genes independently: eight blaKPC, four blaVIM, seven blaNDM-1, and four blaIMP (Figure 3). The similar phenotypic susceptibility pattern of the isolates carrying an independent- versus co-existing ESBL genes were examined. Of the eight isolates bearing the blaKPC gene exclusively, two were phenotypic susceptible to imipenem, and one was susceptible to imipenem plus cefepime. Similarly, independent blaIMP bearing three isolates were sensitive to different cephalosporins plus imipenem. Only one blaNDM-1 carrying one isolate showed sensitivity to imipenem, and the remaining six conferred resistance to all tested β-lactam antibiotics. However, only blaVIM containing four isolates was found to confer complete resistance (Table 3). In contrast, the co-existence of the KPC+VIM, IMP+VIM, NDM-1+IMP genes gave the isolates absolute resistance to the antibiotics tested. Finally, despite co-harboring blaVIM+blaNDM-1, two isolates appeared sensitive to imipenem; however, the other three were completely resistant to all the antibiotics (Table 3).
 |
Table 3 Phenotypic Antibiotic-Resistance of Bacterial Isolates Carrying Single-Type versus Combined-Types ESBLa Genes |
Discussion
This study identified Klebsiella pneumoniae as a prevalent etiology of UTI in Bangladesh, as stated earlier.39 More than half of the K. pneumoniae isolates were found to carry at least one MBL or blaKPC gene. These findings highlighted that the Indian subcontinent countries, including Bangladesh, are a high endemic region for MBL genes among K. pneumoniae and other Enterobacteriaceae.16,40 As a consequence, the evolution of blaKPC- and MBLs-based resistance has been disseminated worldwide in most Gram-negative bacteria.24,41 Additional research studies are needed to develop a deeper understanding of MBL prevalence in other UTI pathogens besides K. pneumoniae in Bangladesh. As anticipated, single occurrences of the blaKPC, blaIMP, blaVIM, and blaNDM-1 genes are associated with a reduced susceptibility to all the β-lactam antibiotics tested regardless of gender and age variation. These findings are supported by many earlier studies.12,13
Further, a co-existence of the blaKPC and MBL genes shows more reduced susceptibility to β-lactam antibiotics. This co-resistance phenomenon enables bacteria to accumulate several genes simultaneously to emerge as high-risk UTI clones. Multiple studies have found that the co-existence of several genes, notably blaKPC and MBLs, into the same bacteria affects decreased sensitivity to different antimicrobials.42,43 Historically, uncomplicated UTIs have been treated with empirical antibiotics, including the cephalosporins, TMP-SMX, and the fluoroquinolones. This widespread use may have led to bacteria acquiring increased co-resistance over time. In Bangladesh, UTI is a frequent disorder at the community level where many patients do not visit medical doctors because of social stigma about issues related to genito-urinary conditions. Therefore, the more common antibiotics are used, which also can lead to resistance. A lack of education, ignorance, and lack of money to buy a full course of an appropriate drug are other factors that can promote high ARG transmission among UTI pathogens in the country.
On the other hand, a significant portion of isolates not carrying those ESBL genes also showed phenotypic resistance to the same sets of β-lactam antibiotics. The inconsistency of the genotype-phenotype association could be explained by other resistance genes or factors that have not been investigated in this study.44 Varieties of ESBLs genes out of KPC or MBL lineage may have contributed to the different phenotypic resistance phenomena.45,46
Uncommonly, this study identified that some K. pneumoniae isolates appeared phenotypic susceptible, particularly to imipenem, in the presence of blaKPC or either of the three MBL genes. Two independent experiments generated the replicated imipenem-sensitive phenotypes of the isolates carrying blaKPC or MBL genes. This indicates that the acquisition of either of the blaKPC, blaIMP, blaVIM or the blaNDM-1 gene is not enough to provide carbapenem-resistance. These findings differed from many other reports stating MBL genes confer resistance to carbapenem-group antibiotics.5,16,17,47 The disparity of ARGs to phenotypic attestation has identified the limitation of molecular assays for antimicrobial resistance determinants. The findings also authenticate the conventional antimicrobial phenotypic testing procedures, notably, disc diffusion, microdilution, and Etest as “gold-standard” methods. The explanation for the expression-discrepancy of the dysfunctional ESBL resistance genes is not well-understood. It is possible that the antibiotic itself may modulate the ARGs into low in vitro expression,48 or that the heteroresistance phenomena associated with unstable tandem gene amplification, rare mutation, and environmental modulation to the resistant genes may explain exhibited different susceptibilities.49–51 Comprehensive whole-genome sequencing (WGS)-based analysis could help to identify accurate genotype to phenotype resistance association.
This study diagnosed UTI more commonly in females than in males. Anatomical differences are reported to affect prevalence variation among the two genders.52 Reproductively active women in the age group 20–39 years accounted for the majority of UTI presentations in this study.53 However, the detection frequency of K. pneumonia in all the age groups was almost equal. Overall, all generations of cephalosporin antibiotics showed a very weak antibacterial potency against the UTI K. pneumoniae tested.53 Higher resistance to these antibiotics may be due to their extensive use in treating UTI and other diseases.54 With the reduced potency of cephalosporins, clinicians have started relying more on carbapenems. This study identified imipenem as the best effective β-lactam antibiotic to treat UTI, although about three-quarters of isolates were resistant to it. A previous study conducted in Bangladesh identified about 15% of imipenem-resistant gram-negative UTI pathogens,40 indicating an increasing trend of carbapenem resistance in uropathogens circulating in the country. Again, the emergence of ESBL or carbapenemase-producing K. pneumoniae can be linked with increased carbapenem consumption.55 The widely used fluoroquinolone drug, ciprofloxacin, remained more effective than any of the β-lactam antibiotics tested under this investigation.
This study has several crucial limitations. The risk behavior data relating to the study participants were self-reported and not evaluated in-depth. This study was cross-sectional in design and follow up could not be carried out due to resource limitations. The convenience sampling was undertaken in a semi-urban area of Bangladesh, where a high degree of social stigma exists about urogenital diseases. Notably, women feel shy or uncomfortable visiting doctors/diagnostics for genitourinary diseases until symptoms become very severe. Therefore, the patients in this study were presenting with acute symptoms and were diagnosed with a higher frequency of positive cases than those in other similar reports.23 This study analyzed only a few ESBL genes and limited β-lactam antibiotics. Only K. pneumoniae were analyzed in the study, despite the existence of diverse other UTI pathogens. This study did not have ATCC-resistant control for AST measurement. We used a P. aeruginosa isolated from a skin wound as an internal resistant-control. The isolate was tested previously resistant against 17 different antibiotics from 7 groups. The small sample size was also a constraint to performing fully powered statistical analyses. However, our results were generated from a resource-limited setting and maintained internal validity by repeating independent experiments where necessary. The findings have shed some light on a significant UTI pathogen, their carriage of ESBL genes, co-resistance, and the genotypic-phenotypic association with antibiotic resistance. Further studies may be required to assess the external validity of our findings.
Conclusions
K. pneumoniae remains a predominant uropathogen in Bangladesh. Imipenem was found the drug of choice among tested β-lactam antibiotics, superior to trimethoprim-sulfamethoxazole and inferior to ciprofloxacin, to treat K. pneumoniae causing UTI. The prominent ESBL genes, namely, blaKPC, blaIMP, blaVIM, and blaNDM-1, emerged in the UTI isolates. A combination of the ESBL genes exerted more reduced phenotypic susceptibility than that of each individual gene. The presence of the four ESBL genes, singly or in conjunction, do not confer absolute resistance to carbapenem antibiotics. The discrepancy of genotype and phenotype resistance has significant consequences for clinical bacteriology, precision in diagnosis, the prudent selection of antimicrobials, and rational prescribing. It highlights that more understanding of antimicrobial resistance is required so that more appropriate and thus, more effective treatments can be provided.
Recommendations
- Diagnostic or clinical laboratories should consider the limitations of molecular methods of identifying antimicrobial resistance.
- Results from adopted molecular techniques should be validated further by phenotypic outcomes. Any discordant results should be repeated by additional testing as per the guidelines of a reference laboratory.
- Heterogeneous phenotypes of antimicrobial susceptibility testing should be taken seriously to avoid imprudent therapeutic decisions.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bush K, Bradford PA. Beta-lactams and beta-lactamase inhibitors: an overview. Cold Spring Harb Perspect Med. 2016;6(8):a025247. doi:10.1101/cshperspect.a025247
2. Knott-Hunziker V, Waley SG, Orlek BS, Sammes PG. Penicillinase active sites: labelling of serine-44 in beta-lactamase I by 6beta-bromopenicillanic acid. FEBS Lett. 1979;99(1):59–61. doi:10.1016/0014-5793(79)80248-3
3. Zhang H, Hao Q. Crystal structure of NDM-1 reveals a common beta-lactam hydrolysis mechanism. FASEB J. 2011;25(8):2574–2582. doi:10.1096/fj.11-184036
4. Ambler RP. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980;289(1036):321–331.
5. Sacha P, Ostas A, Jaworowska J, et al. The KPC type beta-lactamases: new enzymes that confer resistance to carbapenems in Gram-negative bacilli. Folia Histochem Cytobiol. 2009;47(4):537–543. doi:10.2478/v10042-009-0079-y
6. Bush K. Past and present perspectives on beta-lactamases. Antimicrob Agents Chemother. 2018;62(10). doi:10.1128/AAC.01076-18
7. Tehrani K, Martin NI. Beta-lactam/beta-lactamase inhibitor combinations: an update. Medchemcomm. 2018;9(9):1439–1456. doi:10.1039/C8MD00342D
8. Powers RA. Structural and functional aspects of extended-spectrum AmpC cephalosporinases. Curr Drug Targets. 2016;17(9):1051–1060. doi:10.2174/1573399811666150615144707
9. Kotsakis SD, Caselli E, Tzouvelekis LS, Petinaki E, Prati F, Miriagou V. Interactions of oximino-substituted boronic acids and beta-lactams with the CMY-2-derived extended-spectrum cephalosporinases CMY-30 and CMY-42. Antimicrob Agents Chemother. 2013;57(2):968–976. doi:10.1128/AAC.01620-12
10. Docquier JD, Mangani S. Structure-function relationships of class D carbapenemases. Curr Drug Targets. 2016;17(9):1061–1071. doi:10.2174/1389450116666150825115824
11. Walther-Rasmussen J, Hoiby N. OXA-type carbapenemases. J Antimicrob Chemother. 2006;57(3):373–383. doi:10.1093/jac/dki482
12. Emeraud C, Escaut L, Boucly A, et al. Aztreonam plus clavulanate, tazobactam, or avibactam for treatment of infections caused by metallo-beta-lactamase-producing gram-negative bacteria. Antimicrob Agents Chemother. 2019;63(5). doi:10.1128/AAC.00010-19.
13. Wiegand I, Hilpert K, Hancock RE. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008;3(2):163–175. doi:10.1038/nprot.2007.521
14. Gibb AP, Tribuddharat C, Moore RA, et al. Nosocomial outbreak of carbapenem-resistant Pseudomonas aeruginosa with a new bla(IMP) allele, bla(IMP-7). Antimicrob Agents Chemother. 2002;46(1):255–258. doi:10.1128/AAC.46.1.255-258.2002
15. Yong D, Toleman MA, Giske CG, et al. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53(12):5046–5054. doi:10.1128/AAC.00774-09
16. Kumarasamy KK, Toleman MA, Walsh TR, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10(9):597–602. doi:10.1016/S1473-3099(10)70143-2
17. Cornaglia G, Giamarellou H, Rossolini GM. Metallo-beta-lactamases: a last frontier for beta-lactams? Lancet Infect Dis. 2011;11(5):381–393. doi:10.1016/S1473-3099(11)70056-1
18. Pournaras S, Poulou A, Voulgari E, Vrioni G, Kristo I, Tsakris A. Detection of the new metallo-beta-lactamase VIM-19 along with KPC-2, CMY-2 and CTX-M-15 in Klebsiella pneumoniae. J Antimicrob Chemother. 2010;65(8):1604–1607. doi:10.1093/jac/dkq190
19. Willyard C. The drug-resistant bacteria that pose the greatest health threats. Nature. 2017;543(7643):15. doi:10.1038/nature.2017.21550
20. Friedman ND, Carmeli Y, Walton AL, Schwaber MJ. Carbapenem-resistant enterobacteriaceae: a strategic roadmap for infection control. Infect Control Hosp Epidemiol. 2017;38(5):580–594. doi:10.1017/ice.2017.42
21. Rodriguez-Bano J, Gutierrez-Gutierrez B, Machuca I, Pascual A. Treatment of infections caused by extended-spectrum-beta-lactamase-, AmpC-, and carbapenemase-producing enterobacteriaceae. Clin Microbiol Rev. 2018;31(2).
22. Kelly AM, Mathema B, Larson EL. Carbapenem-resistant Enterobacteriaceae in the community: a scoping review. Int J Antimicrob Agents. 2017;50(2):127–134. doi:10.1016/j.ijantimicag.2017.03.012
23. Arana DM, Rubio M, Alos JI. Evolution of antibiotic multiresistance in Escherichia coli and Klebsiella pneumoniae isolates from urinary tract infections: a 12-year analysis (2003–2014). Enferm Infecc Microbiol Clin. 2017;35(5):293–298. doi:10.1016/j.eimc.2016.02.018
24. Logan LK, Weinstein RA. The epidemiology of carbapenem-resistant enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis. 2017;215(suppl_1):S28–S36. doi:10.1093/infdis/jiw282
25. Melgarejo JL, Cardoso MH, Pinto IB, et al. Identification, molecular characterization, and structural analysis of the blaNDM-1 gene/enzyme from NDM-1-producing Klebsiella pneumoniae isolates. J Antibiot (Tokyo). 2019;72(3):155–163. doi:10.1038/s41429-018-0126-z
26. Balm MN, La MV, Krishnan P, Jureen R, Lin RT, Teo JW. Emergence of Klebsiella pneumoniae co-producing NDM-type and OXA-181 carbapenemases. Clin Microbiol Infect. 2013;19(9):E421–423. doi:10.1111/1469-0691.12247
27. Davis MA, Besser TE, Orfe LH, et al. Genotypic-phenotypic discrepancies between antibiotic resistance characteristics of Escherichia coli isolates from calves in management settings with high and low antibiotic use. Appl Environ Microbiol. 2011;77(10):3293–3299. doi:10.1128/AEM.02588-10
28. Pink RC, Wicks K, Caley DP, Punch EK, Jacobs L, Carter DR. Pseudogenes: pseudo-functional or key regulators in health and disease? RNA. 2011;17(5):792–798. doi:10.1261/rna.2658311
29. Wilson ML, Gaido L. Laboratory diagnosis of urinary tract infections in adult patients. Clin Infect Dis. 2004;38(8):1150–1158. doi:10.1086/383029
30. Stamm WE, Counts GW, Running KR, Fihn S, Turck M, Holmes KK. Diagnosis of coliform infection in acutely dysuric women. N Engl J Med. 1982;307(8):463–468. doi:10.1056/NEJM198208193070802
31. Odoki M, Almustapha Aliero A, Tibyangye J, et al. Prevalence of bacterial urinary tract infections and associated factors among patients attending hospitals in Bushenyi District, Uganda. Int J Microbiol. 2019;2019:4246780. doi:10.1155/2019/4246780
32. van der Zee A, Roorda L, Bosman G, Ossewaarde JM, Lin B. Molecular diagnosis of urinary tract infections by semi-quantitative detection of uropathogens in a routine clinical hospital setting. PLoS One. 2016;11(3):e0150755. doi:10.1371/journal.pone.0150755
33. CLSI. Performance Standards for Antimicrobial Susceptibility Testing.
34. Institute. CaLS. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically.
35. Papp-Wallace KM, Endimiani A, Taracila MA, Bonomo RA. Carbapenems: past, present, and future. Antimicrob Agents Chemother. 2011;55(11):4943–4960. doi:10.1128/AAC.00296-11
36. Testing. ECoAS. Breakpoint tables for interpretation of MICs and zone diameters. EUCAST. 2017;Version 7.1.
37. Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119–123. doi:10.1016/j.diagmicrobio.2010.12.002
38. Senda K, Arakawa Y, Ichiyama S, et al. PCR detection of metallo-beta-lactamase gene (blaIMP) in gram-negative rods resistant to broad-spectrum beta-lactams. J Clin Microbiol. 1996;34(12):2909–2913. doi:10.1128/JCM.34.12.2909-2913.1996
39. Hooton TM. Pathogenesis of urinary tract infections: an update. J Antimicrob Chemother. 2000;46(90001):1–7. doi:10.1093/jac/46.suppl_1.1
40. Begum N, Shamsuzzaman SM. Emergence of carbapenemase-producing urinary isolates at a tertiary care hospital in Dhaka, Bangladesh. Ci Ji Yi Xue Za Zhi. 2016;28(3):94–98. doi:10.1016/j.tcmj.2016.04.005
41. Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis. 2009;9(4):228–236. doi:10.1016/S1473-3099(09)70054-4
42. Chen Z, Qlu S, Wang Y, et al. Coexistence of blaNDM-1 with the prevalent blaOXA23 and blaIMP in pan-drug resistant Acinetobacter baumannii isolates in China. Clin Infect Dis. 2011;52(5):692–693. doi:10.1093/cid/ciq231
43. Canton R, Ruiz-Garbajosa P. Co-resistance: an opportunity for the bacteria and resistance genes. Curr Opin Pharmacol. 2011;11(5):477–485. doi:10.1016/j.coph.2011.07.007
44. Bengtsson-Palme J, Kristiansson E, Larsson DGJ. Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol Rev. 2018;42(1). doi:10.1093/femsre/fux053
45. Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev. 2005;18(4):657–686. doi:10.1128/CMR.18.4.657-686.2005
46. Bradford PA. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. 2001;14(4):933–951, table of contents. doi:10.1128/CMR.14.4.933-951.2001
47. Naas T, Dortet L, Iorga BI. Structural and functional aspects of class A carbapenemases. Curr Drug Targets. 2016;17(9):1006–1028. doi:10.2174/1389450117666160310144501
48. Depardieu F, Podglajen I, Leclercq R, Collatz E, Courvalin P. Modes and modulations of antibiotic resistance gene expression. Clin Microbiol Rev. 2007;20(1):79–114. doi:10.1128/CMR.00015-06
49. Andersson DI, Nicoloff H, Hjort K. Mechanisms and clinical relevance of bacterial heteroresistance. Nat Rev Microbiol. 2019;17(8):479–496. doi:10.1038/s41579-019-0218-1
50. Nicoloff H, Hjort K, Levin BR, Andersson DI. The high prevalence of antibiotic heteroresistance in pathogenic bacteria is mainly caused by gene amplification. Nat Microbiol. 2019;4(3):504–514. doi:10.1038/s41564-018-0342-0
51. Hughes D, Andersson DI. Environmental and genetic modulation of the phenotypic expression of antibiotic resistance. FEMS Microbiol Rev. 2017;41(3):374–391. doi:10.1093/femsre/fux004
52. Minardi D, d’Anzeo G, Cantoro D, Conti A, Muzzonigro G. Urinary tract infections in women: etiology and treatment options. Int J Gen Med. 2011;4:333–343. doi:10.2147/IJGM.S11767
53. Akoachere JF, Yvonne S, Akum NH, Seraphine EN. Etiologic profile and antimicrobial susceptibility of community-acquired urinary tract infection in two Cameroonian towns. BMC Res Notes. 2012;5:219. doi:10.1186/1756-0500-5-219
54. Meyer E, Schwab F, Schroeren-Boersch B, Gastmeier P. Dramatic increase of third-generation cephalosporin-resistant E. coli in German intensive care units: secular trends in antibiotic drug use and bacterial resistance, 2001 to 2008. Crit Care. 2010;14(3):R113. doi:10.1186/cc9062
55. Klein EY, Van Boeckel TP, Martinez EM, et al. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc Natl Acad Sci U S A. 2018;115(15):E3463–E3470. doi:10.1073/pnas.1717295115
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.