Back to Journals » Clinical, Cosmetic and Investigational Dermatology » Volume 17
Genomics-Microbiome Based Assessment of Bidirectional Causality Between Gut Microbiota and Psoriasis
Authors Gao Q , Liu JH, Ma WY , Cheng ZL, Hao PS , Luo NN
Received 19 November 2023
Accepted for publication 8 January 2024
Published 13 February 2024 Volume 2024:17 Pages 435—445
DOI https://doi.org/10.2147/CCID.S450227
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Anne-Claire Fougerousse
Qian Gao,1,2 Jing-Hua Liu,1,2 Wen-Yi Ma,1,2 Zi-Lin Cheng,1,2 Ping-Sheng Hao,1 Na-Na Luo1
1Hospital of Chengdu University of Traditional Chinese Medicine, Chengdu, People’s Republic of China; 2Chengdu University of Traditional Chinese Medicine, Chengdu, People’s Republic of China
Correspondence: Ping-Sheng Hao; Na-Na Luo, Email [email protected]; [email protected]
Background: Traditional observational studies have found a possible risk association of the gut microbiota for psoriasis. Meanwhile, psoriasis may also affect the changes in the gut microbiota. However, the available evidence does not demonstrate a reciprocal relationship between the gut microbiota and psoriasis. This limits our understanding on the role of the gut microbiota in the mechanisms of psoriasis.
Methods: To address this question we used Mendelian randomization, a novel epidemiological approach, and acquired the largest current gut microbiota GWAS data from the MiBioGen consortium as well as psoriasis GWAS data from the FinnGen consortium, and performed two-sample bidirectional MR analyses using a multiple MR analysis approach. Finally, the robustness of the results was assessed by sensitivity analysis.
Results: Our results indicate that five bacterial genera are causally related to psoriasis and psoriasis is causally related to four bacterial genera.
Conclusion: These results suggest a bidirectional causal influence of psoriasis on the gut microbiota. Our results somewhat challenge the causal inferences of previous observational studies. We found that the specific bacterial genera with a risk effect on psoriasis were different from those found to characterize psoriasis in previous observational studies, and that these psoriasis-characterizing genera were inversely associated with psoriasis.
Keywords: Mendelian randomization, psoriasis, causal relationship, gut microbiota, bidirectional Mendelian randomization analysis
Introduction
Psoriasis is a chronic and complex skin disease with systemic multisystem comorbidities that occurs in approximately 2.4% of the world’s population1. Current treatments for psoriasis have limited effectiveness and are prone to relapse after discontinuation of medication, which seriously affects the patient’s survival rate and quality of life. Recently, WHO has proposed psoriasis as a major global health problem.2 Psoriasis pathogenesis mainly involves immunity, genetic susceptibility, and environmental factors.3 Among them, the role of intestinal microbiota in psoriasis pathogenesis has received increasing attention. Many studies have shown that changes in the intestinal microbiota may play a non-negligible role in psoriasis. For example, epidemiologic investigations have found an increased risk of psoriasis in inflammatory bowel disease, obesity,4 and other conditions accompanied by intestinal dysbiosis. There are also several observational studies that have shown by macro-genetic sequencing of fecal samples that the composition of the intestinal flora is significantly different between psoriasis patients and healthy controls.5 Not only that, but patients with psoriasis also have significant dysregulation of intestinal microbiota metabolites, such as fatty acid binding protein and tight junction protein-3.6 In addition, probiotics significantly improved skin lesion severity in psoriasis-like mice.7 This evidence suggests that the gut microbiota may be strongly associated with psoriasis, but it is not clear whether changes in gut flora trigger psoriasis or whether psoriasis causes changes in gut flora. In addition, it should not be overlooked that observational studies are susceptible to confounding factors leading to abnormally skewed results, such as metabolic disorders, cardiovascular disease, and obesity (as a psoriasis comorbidity), as well as age, dietary variations, and antibiotic use are all independent factors affecting the gut microbiota8–10. The findings of the above observational studies limit the ability to make causal inferences about the gut microbiota and psoriasis. Therefore high-quality studies are necessary to determine the direct causal effect of gut microbiota and psoriasis.
Mendelian randomization (MR) is a latest epidemiological research method that is based on Mendelian laws of inheritance and uses genetic variation as an instrumental variable for causal inference. Unlike traditional randomized controlled trials (RCTs), MR analyses are not influenced by human factors and can minimize bias anomalies caused by confounding factors.11,12 Current large-scale genome-wide association studies (GWAS) provide extensive statistical data for Mendelian randomization studies. To date, Mendelian randomization studies have been successfully applied to explore the causal impact of potential risk factors with complex diseases.13–15
Given the strengths of MR, we performed a comprehensive two-sample bidirectional causal analysis using gut microbiota and psoriasis GWAS data from the public databases MiBioGen Consortium and FinnGen Consortium, respectively, and referenced the STROBE-MR checklist (https://www.STROBE-MR.org/) to ensure the quality and accuracy of the study16. In this study, we sought to elucidate whether the gut flora has a causal role in psoriasis and whether psoriasis has a potential impact on the gut microbiome. This will provide a research basis for the mechanisms of the intestinal microbiota’s role in psoriasis and provide new biomarkers and therapeutic strategies by characterizing causal effects between specific genera of bacteria and psoriasis patients.
Materials and Methods
Research Design
Figure 1 shows our overall study design for MR. Our design maximizes the possibility of avoiding the influence of confounding factors. Here, we first acquired independent GWAS data on gut microbiota and psoriasis from different databases. In addition, we implemented the MR study to fulfill the following three key assumptions (Figure 2): (1) SNPs of instrumental variables (IVs) are strongly associated with exposure; (2) SNPs are not associated with any confounders of exposure and outcome associations; (3) SNPs act only through exposure and not through other pathways.17 To fulfill the above assumptions we screened candidate instrumental variables after a series of criteria and used five MR analyses to ensure the robustness of the results. Finally, the results were tested by sensitivity analysis in order to detect the possibility of heterogeneity and horizontal pleiotropy. Then, it was followed by reverse MR analysis.
 |
Figure 1 A schematic of the design and process of our MR study. |
Data Sources
The gut microbiota GWAS data were from the MiBioGen consortium (https://mibiogen.gcc.rug.nl/). This is the largest multi-ethnic genome-wide association meta-analysis of gut microbiota to date, harmonizing 18,340 individuals from 24 cohorts of different countries, ages, and genders.18 Genome-wide genotypes were obtained by targeted 16S rRNA gene sequencing of fecal samples (using seven different fecal extraction methods and three different 16S rRNA regions) for a total of 211 taxa (131 genus, 35 family, 20 order, 16 class, and 9 phylum). This GWAS data is now widely used in MR studies of the gut microbiota.19–21
Psoriasis GWAS data were obtained from the FinnGen database. FinnGen study is a continuously updated comprehensive database containing genetic information data and health registry records (https://www.FinnGen.fi/en). We chose the most recent and larger sample sizes data to ensure the statistical efficacy of our study. The psoriasis data we retrieved (finn-b-L12_PSORIASIS) were coded using ICD-10-L40 and included a total of 4510 cases and 212,242 controls, all from the European population.22
Instrumental Variables Selection
We set the following steps to screen the best IVs to ensure the robustness of our study; first, screening of significantly relevant SNPs from gut microbiome GWAS data as IVs. However there is a problem that is prevalent in MR studies of the gut microbiota19,23. If the number of SNPs obtained by using the genome-wide statistical significance threshold (p<5×10-8) is few, it may lead to biased results. In order to obtain more convincing results, we referred to more MR studies of the gut microbiota and finally used a looser level of genome-wide significance (p<1×10-5).24,25 Second, on the basis of the SNPs obtained in the previous step, we set the linkage disequilibrium filtering conditions to R2 = 0.01 and clumping distance = 10,000 kb. Such conditions ensure statistical efficacy while minimizing the risk of confounding.26 Third, removal of palindromic SNPs with A/T or G/C structures from SNPs obtained in the second step.19,26 Finally, we calculated the F statistic to detect the strength of the correlation between the SNPs obtained in the third step and the exposure, removing SNPs with weak instrumental bias (F values <10).27
Mendelian Randomization Analysis
This study was conducted in R software (4.3.0), the main R packages we used included TwoSampleMR, MR-PRESSO. When there was only one IV in the exposure results, causality was tested using the Wald ratio method. When there were multiple IVs, we used five different MR analysis methods; inverse variance weighted (IVW), MR-Egger regression, weighted median, weighted mode, and simple mode. In the absence of pleiotropy, inverse variance weighting (IVW) is the most reliable analysis method. IVW has high accuracy because it combines the ratio of each SNP to estimate the causal effect of exposure on outcome.27 The other four methods have their own focus and are often used as complementary methods to ensure more robust results. Weighted median estimates are robust to the presence of null instruments and provide consistent estimates of causal effects even when up to 50% of the instrumental variables are null.24 Weighted models remain valid even when other instrumental variables do not satisfy MR requirements. MR-Egger not only assesses the presence of pleiotropy in genetic variation, but also provides estimates when pleiotropy is present.28
Sensitivity Analysis
We tested for sensitivity using a variety of methods. The distribution of the funnel plot is used to show whether the results are biased or not. Leave-One-Out (LOO) analysis detects whether the removal of a single SNP affects the direction of the overall causal association.28 MR-Egger intercept detects the presence of pleiotropy by evaluating the distance of the intercept from zero. MR-PRESO is a more robust method for detecting pleiotropy and reduces horizontal pleiotropy by removing outliers.29 Cochran’ s Q statistic was used to detect heterogeneity among the selected genetic variants, and the effect of heterogeneity should be considered if P<0.05.
Results
Causal Association of Gut Microbiota on Psoriasis
Based on the screening criteria we set, a total of 2876 SNPs from 211 bacterial genera were selected as candidate IVs, respectively, at the phylum (124), class (229), order (286), family (505), and genus (1732). The F-values of these SNPs were all >10, which suggests a strong correlation with the gut microbiota (Supplementary Table S1).
Full results of the preliminary analysis showing the relationship between gut microbiota and psoriasis risk are provided in Figure 3 and Supplementary Table S2. We found four genera to be associated with psoriasis, specifically Eubacteriumfissicatenagroup, Lactococcus, Odoribacter, and Ruminiclostridium5. At least one of these MR methods was found to be associated with psoriasis (Table 1; Supplementary Figure S1). Our results showed a negative effect of Eubacteriumfissicatenagroup on psoriasis (IVW OR 1.27; 95% CI 1.1–1.47; p=1.10×10-3), and also supported by a Weighted median (OR 1.27; 95% CI 1.04–1.54; p=1.7×10-2). However, it is worth noting that the MR-Egger analysis produced effect estimates in the opposite direction from the other four MR analyses (IVW, weighted model, weighted median, and simple model), albeit not significantly. This situation exists in other MR studies as well.23 More stringent conditions should be set to screen IVs to repeat the analyses, such as narrowing the genome-wide significance threshold, but more stringent screening conditions would result in very few selected SNPs, which would make the results unconvincing. Considering that IVW estimates are more precise than MR-Egger in the absence of horizontal pleiotropy, and that MR-Egger is susceptible to peripheral genetic variables, the results of the analysis for Eubacteriumfissicatenagroup are acceptable, but more studies are still needed to argue the case. Similarly, Lactococcus had a negative impact on psoriasis (IVW OR 1.25; 95% CI 1.08–1.44; p=2.80×10-3), and the presence of an MR-Egger estimated effect in the reverse direction of the results of the other four MR analyses. For such a result we choose to be cautious in our reservations. Ruminiclostridium5 also had a negative influence on psoriasis (IVW OR 1.31; 95% CI 1.01–1.7; p=4.3×10-2) and in the same direction as the results of the other four MR analyses. In addition, our findings showed Odoribacter suggested a protective effect against psoriasis (IVW OR 0.71; 95% CI 0.53–0.96; p=2.40×10-2), and were in the same direction as the results of the other four MR analyses.
 |
Table 1 MR Estimates for the Causal Association of Psoriasis on Four Gut Microbiota |
For these four potential causal relationships we conducted sensitivity analyses, and the results of the MR-Egger intercept and Cochran’s Q showed no indication of the presence of horizontal pleiotropy and heterogeneity (p>0.05) (Table 2). Most funnel plots were symmetrical, except for Lactococcus (Supplementary Figure S2). In the leave-one-out analysis, there were aberrant IVs in Odoribacter and Ruminiclostridium5 (Supplementary Figure S3). However, more robust MR-PRESO did not detect the presence of abnormal IVs (Table 2).
 |
Table 2 Sensitivity Analysis of the Causal Association Between Four Gut Microbiota and Psoriasis |
Causal Association of Psoriasis on Gut Microbiota
Reverse MR analyses identified a potential causal effect of psoriasis on five genera of enteric bacteria (Table 3 and Supplementary Figure S4), and the complete results of the analyses are in the Supplementary Data (Supplementary Table S3; Supplementary Table S4). Of these, psoriasis had a risk effect on four genera, including Bacteroidia (IVW OR 1.04;95% CI 1–1.07; p = 0.04), Intestinimonas (IVW OR 1.04;95% CI 1.01–1.08; p = 0.02), Bacteroidales (IVW OR 1.04;95% CI 1–1.07; p = 0.04), and Bacteroidetes (IVW OR 1.04; 95% CI 1 −1.07; p = 0.04). Surprisingly, psoriasis had a positive association with gut microbiota, Proteobacteria (IVW OR 0.97;95% CI 0.95–1; p = 0.05). Although the results of the other four analyses were not significant, they were in the same direction as IVW. Potential horizontal pleiotropy and heterogeneity were not shown in the sensitivity analysis (Table 4; Supplementary Figure S5; Supplementary Figure S6).
 |
Table 3 MR Estimates for the Causal Association of Psoriasis on Five Gut Microbiota |
 |
Table 4 Sensitivity Analysis of the Causal Association Between Psoriasis and Five Gut Microbiota |
Discussion
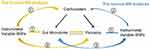
Notably, this is the first report of using MR to investigate the bidirectional causal relationship between psoriasis and gut microbiome (Figure 4). As we used a large sample size of GWAS data and a more robust MR design, we provide convincing evidence for our MR results. We identified four specific genera with a risk effect on psoriasis, in addition, a potential causal effect of psoriasis on five genera. These results establish a bidirectional causal effect between gut microbiota changes and psoriasis. Our results somewhat challenge the causal inferences of previous observational studies. The specific genera we found to have a risk effect on psoriasis were different from the characteristic genera found in observational studies, and the association between these psoriasis-characterizing genera and psoriasis was inverse. Our results will be necessary for further research in this area.
 |
Figure 4 Bidirectional MR results of causal effects between gut microbiome and psoriasis. |
Studies have shown that the intestinal microbiota may play an important role in the pathogenesis of many complex diseases. In light of the proposed “gut-skin axis” and the presence of altered gut microbiota composition and aberrant bacterial metabolites in many inflammatory skin diseases,30 there has been widespread interest in the relevance of the gut microbiota to skin homeostasis. Among them, several observational studies have found significant variability in the gut microbiota of psoriasis patients. It is now agreed that the relative abundance and diversity of intestinal flora organisms are significantly altered in psoriasis patients compared to healthy individuals, with some differences at the phylum, order, genus, and species levels.31–33 However, previous studies with differences in design and methodology have led to different or even conflicting results in assessing changes in genera.34 For example, Firmicutes and Bacteroidetes are the main genera in patients with psoriasis,35 with Firmicutes accounting for about 45% and Bacteroidetes accounting for about 36% of the total.36 Some studies have found an increased Firmicutes/Bacteroidetes ratio in psoriasis patients compared to healthy controls by 16S rRNA sequencing of fecal samples.33,37,38 However, Huang et al reported inconsistent results, found a decreased in Firmicutes and an enrichment in Bacteroidetes in the psoriasis patient group.36 Similarly, Akkermansia is considered to be a “psoriasis characteristic genera”39 however, changes in the abundance of Akkermansia in psoriasis patients have been inconsistently reported.33,40 As the degree of gut dysbiosis in psoriasis patients correlates with lesion severity, inflammation-related markers (IL-2, CRP),31,36 and comorbidities (obesity, IBD). It is difficult to draw consistent conclusions about changes in the intestinal microbiota in psoriasis patients, which will make the search for the key genera affecting psoriasis more difficult.
In comparison with previous studies, our MR study avoids confounding elements from interfering with the results and has new findings results. Our study confirms that intestinal microbiota dysbiosis can trigger the development of psoriasis. Our study found that at the genus level, Eubacteriumfissicatenagroup, Lactococcus, and Odoribacter had a risky effect on psoriasis. Of these, only Lactococcus has been reported in a previous observational study, which indicated that psoriasis patients were relatively more abundant in Lactococcus compared to healthy persons,36 as shown by the Wilcoxon test, which is consistent with our study. Lactococcus has been reported to be associated with high-fat diets and obesity,41,42 which is also a predisposing factor for psoriasis. However, an association between the Eubacteriumfissicatenagroup and Odoribacter with psoriasis has not been reported in any study. Short-chain fatty acids (SCFAs) are metabolites converted by fermentation of otherwise indigestible dietary fiber by intestinal bacteria and are important in maintaining intestinal barrier function and anti-inflammatory properties.43 Eubacterium spp. and Odoribacter are among the known producers of SCFAs, that maintains intestinal homeostasis.44,45 They are currently reported to have a positive role in metabolic syndrome, inflammatory bowel disease.46 However, our study showed that they have a dangerous impact on the developing of psoriasis. Hence, more studies are needed to further substantiate the role of Eubacterium spp. and Odoribacter in psoriasis. In addition, in contrast to previous studies, our study was the first to find a protective effect of Ruminiclostridium5 against psoriasis. Some studies have reported that Ruminiclostridium5 may be associated with glucose tolerance, high-fat diet.47,48 Its effect on the development of psoriasis is unknown.
Reverse MR analysis revealed that psoriasis has a dangerous effect on Bacteroidia, Intestinimonas, Bacteroidetes, and Bacteroidales, which confirms that psoriasis can cause dysbiosis of intestinal genera. Observational studies have shown that Bacteroidetes is a key genus for gut microbiota changes in psoriasis patients.36 Our findings similarly identified a potential association between Bacteroidetes and psoriasis, but this association was reverse causality, which somewhat challenges the causal inference of previous studies. Meanwhile, previous reports on the abundance of Bacteroidetes have been inconsistent, and our results provide illustrative evidence for this point of contention. In addition, our study is the first to report the effect of psoriasis on Intestinimonas, and no study has yet reported linking Intestinimonas to psoriasis. Intestinimonas is a newly described genus of metabolically characterized bacteria with unique metabolic profiles, producing butyrate with anticancer, anti-inflammatory, and antioxidant effects.49 It has been shown to be associated with a high-sugar diet, inflammatory bowel disease.50,51 Our study also found a protective effect of psoriasis on Proteobacteria. However, existing studies found a significant reduction of Proteobacteria in psoriasis patients and an increase in its relative abundance after secukinumab treatment, which contradicts our results. Therefore, more studies are necessary to argue about the effect of psoriasis on Proteobacteria. Our study confirms that the changes in the abundance of Bacteroidia, Bacteroidetes, and Proteobacteria are caused by psoriasis, and laterally verifies that the treatment of psoriasis significantly improves the intestinal microbiota situation, but the concrete mechanism requires further clarification.
Although our study has been adequately designed, there are still some limitations that must be confronted: concerning the data sources, the GWAS data included in this study were primarily from subjects of European ancestry, which does not exclude the existence of overlapping participants, but our application of F-statistics can minimize the bias of overlapping participants.28 Additionally, it is well known that the incidence of psoriasis and the distribution of gut microbiota differ between human races, so the results of this study have limited validity for other races. Some observational studies have shown that variability in gut microbiota correlates with the severity of psoriasis,36 but the available GWAS data do not grade the severity of psoriasis in patients with psoriasis, and therefore fail to assess the effect of genetic variance in the changes in gut microbiota in relation to the grading of the degree of psoriasis. Furthermore, because of the small sample size of the gut microbiome GWAS data, a more moderate GWAS significance threshold (P<1×10-5) was used in our study to enable as many SNPs as possible to be screened for effective MR analysis. Finally, the inconsistency in the direction of the results of MR-Egger and the other four analyses in our results provided a great example of the rigor of using multiple analytical methods, and since the IVW analysis is more robust than MR-Egger, we retained these positive results, which is acceptable for our exploratory study.
Conclusions
In conclusion, our study identifies a bidirectional potential association between specific gut microbiota and psoriasis and discovers potential causative genera of psoriasis. This is positive for a deeper understanding of the role of gut microbiota in psoriasis and could contribute to the development of interventions for gut flora in psoriasis. It should be emphasized that more experiments are necessary to validate these associations.
Data Sharing Statement
Gut microbiome GWAS data can be downloaded for free on https://mibiogen.gcc.rug.nl/. Psoriasis GWAS data can be retrieved on https://risteys.finregistry.fi/endpoints/L12_PSORIASIS.
Ethics Approval and Consent to Participate
The GWAS data applied in our study were obtained from public databases, and the original study was approved by informed consent and ethical review boards of all participants. Therefore, our study could be conducted without additional ethical approval.
Acknowledgments
The GWAS data utilized in this study were obtained from the MiBioGen Consortium and FinnGen databases, and it is undoubtedly due to these participants and researchers that the data they provided for the study were important to our research.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was supported by the Sichuan Provincial Science and Technology Department Foundation (Grant No:2020YJ0436).
Disclosure
The authors declare no conflict of interest.
References
1. Langley RGB. Psoriasis: epidemiology, clinical features, and quality of life. Ann Rheumatic Dis. 2005;64(suppl_2):ii18–ii23. doi:10.1136/ard.2004.033217
2. Boehncke W-H, Schön MP. Psoriasis. Lancet. 2015;386(9997):983–994. doi:10.1016/S0140-6736(14)61909-7
3. Kamiya K, Kishimoto M, Sugai J, et al. Risk factors for the development of psoriasis. Int J Mol Sci. 2019;20(18):4347. doi:10.3390/ijms20184347
4. Jensen P, Skov L. Psoriasis and obesity. Dermatology. 2016;232(6):633–639. doi:10.1159/000455840
5. Langan EA, Künstner A. The gastrointestinal microbiome and psoriasis: more food for thought. Br J Dermatol. 2022;187(2):2. doi:10.1111/bjd.21618
6. Visser MJE, Kell DB, Pretorius E. Bacterial dysbiosis and translocation in psoriasis vulgaris. Front Cell Infect Microbiol. 2019;2019:9.
7. Atabati H, Esmaeili S, Saburi E, et al. Probiotics with ameliorating effects on the severity of skin inflammation in psoriasis: evidence from experimental and clinical studies. J Cell Physiol. 2020;235(12):8925–8937. doi:10.1002/jcp.29737
8. Gagnon E, Mitchell PL, Manikpurage HD, et al. Impact of the gut microbiota and associated metabolites on cardiometabolic traits, chronic diseases and human longevity: a Mendelian randomization study. J Transl Med. 2023;21:1.
9. Sanna S, van Zuydam NR, Mahajan A, et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat Genet. 2019;51(4):600–605. doi:10.1038/s41588-019-0350-x
10. Liu B, Ye D, Yang H, et al. Two-sample Mendelian randomization analysis investigates causal associations between gut microbial genera and inflammatory bowel disease, and specificity causal associations in ulcerative colitis or crohn’s disease. Front Immunol. 2022;2022:13.
11. Sekula P, Del Greco M MFDG, Pattaro C, et al. Mendelian randomization as an approach to assess causality using observational data. J Am Soc Nephrol. 2016;27(11):3253–3265. doi:10.1681/ASN.2016010098
12. Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol. 2017;46(6):1734–1739. doi:10.1093/ije/dyx034
13. Carter P, Yuan S, Kar S, et al. Coffee consumption and cancer risk: a Mendelian randomisation study. Clin Nutr. 2022;41(10):2113–2123. doi:10.1016/j.clnu.2022.08.019
14. Xu J, Zhang X, Tong W, et al. Phenome-wide Mendelian randomization study evaluating the association of circulating vitamin D with complex diseases. Frontiers in Nutrition. 2023;10:1.
15. Kuppa A, Tripathi H, Al-Darraji A, et al. C-reactive protein levels and risk of cardiovascular diseases: a two-sample bidirectional Mendelian randomization study. Int J Mol Sci. 2023;24(11):9129. doi:10.3390/ijms24119129
16. Skrivankova VW, Richmond RC, Woolf BAR, et al. Strengthening the reporting of observational studies in epidemiology using Mendelian randomisation (STROBE-MR): explanation and elaboration. BMJ. 2021:n2233. doi:10.1136/bmj.n2233
17. Yeung SLA, Gill D. Standardizing the reporting of Mendelian randomization studies. BMC Med. 2023;21(1):1. doi:10.1186/s12916-023-02894-8
18. Kurilshikov A, Medina-Gomez C, Bacigalupe R, et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat Genet. 2021;53(2):156–165. doi:10.1038/s41588-020-00763-1
19. Shang W, Zhang S, Qian H, et al. Association of gut microbiota with COVID‐19 susceptibility and severity: a two‐sample Mendelian randomization study. J med virol. 2023;95:4. doi:10.1002/jmv.28734
20. Liu K, Cai Y, Song K, et al. Clarifying the effect of gut microbiota on allergic conjunctivitis risk is instrumental for predictive, preventive, and personalized medicine: a Mendelian randomization analysis. EPMA J. 2023;14(2):235–248. doi:10.1007/s13167-023-00321-9
21. Song J, Wu Y, Yin X, et al. The causal links between gut microbiota and COVID‐19: a Mendelian randomization study. J med virol. 2023;95(5):5. doi:10.1002/jmv.28784
22. Kurki MI, Karjalainen J, Palta P, et al. FinnGen: unique genetic insights from combining isolated population and national health register data. MedRxiv. 2022;2022:1.
23. Luo M, Cai J, Luo S, et al. Causal effects of gut microbiota on the risk of chronic kidney disease: a Mendelian randomization study. Front Cell Infect Microbiol. 2023;13:1.
24. Li C, Liu C, Li N. Causal associations between gut microbiota and adverse pregnancy outcomes: a two-sample Mendelian randomization study. Front Microbiol. 2022;13:1.
25. Zhuang Z, Yang R, Wang W, et al. Associations between gut microbiota and Alzheimer’s disease, major depressive disorder, and schizophrenia. J Neuroinflammation. 2020;17(1). doi:10.1186/s12974-020-01961-8
26. Long Y, Tang L, Zhou Y, et al. Causal relationship between gut microbiota and cancers: a two-sample Mendelian randomisation study. BMC Med. 2023;21(1). doi:10.1186/s12916-023-02761-6
27. Luo S, Li W, Li Q, et al. Causal effects of gut microbiota on the risk of periodontitis: a two-sample Mendelian randomization study. Front Cell Infect Microbiol. 2023;13:1.
28. Xiang K, Wang P, Xu Z, et al. Causal effects of gut microbiome on systemic lupus erythematosus: a two-sample Mendelian randomization study. Front Immunol. 2021;12:1.
29. Zhang L, Wang Y, Qiu L, et al. Psoriasis and cardiovascular disease risk in European and East Asian populations: evidence from meta-analysis and Mendelian randomization analysis. BMC Med. 2022;20(1). doi:10.1186/s12916-022-02617-5
30. Stec A, Sikora M, Maciejewska M, et al. Bacterial metabolites: a link between gut microbiota and dermatological diseases. Int J Mol Sci. 2023;24(4):3494. doi:10.3390/ijms24043494
31. Buhaș MC, Gavrilaș L, Candrea R, et al. Gut microbiota in psoriasis. Nutrients. 2022;14(14):2970. doi:10.3390/nu14142970
32. Wang X, Zhai W, Ma J, et al. Substantial alterations of the intestinal microbiota in psoriasis patients of China. Exp Dermatol. 2021;30(12):1840–1841. doi:10.1111/exd.14295
33. Hidalgo‐Cantabrana C, Gómez J, Delgado S, et al. Gut microbiota dysbiosis in a cohort of patients with psoriasis. Br J Dermatol. 2019;181(6):1287–1295. doi:10.1111/bjd.17931
34. Scher JU, Ubeda C, Artacho A, et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol. 2014;67(1):128–139. doi:10.1002/art.38892
35. Hou K, Wu Z-X, Chen X-Y, et al. Microbiota in health and diseases. Signal Transduct Target Ther. 2022;7(1):1.
36. Huang L, Gao R, Yu N, et al. Dysbiosis of gut microbiota was closely associated with psoriasis. Sci China Life Sci. 2019;62(6):807–815. doi:10.1007/s11427-018-9376-6
37. Chen Y, Ho HJ, Tseng C, et al. Intestinal microbiota profiling and predicted metabolic dysregulation in psoriasis patients. Exp Dermatol. 2018;27(12):1336–1343. doi:10.1111/exd.13786
38. Shapiro J, Cohen NA, Shalev V, et al. Psoriatic patients have a distinct structural and functional fecal microbiota compared with controls. J Dermatol. 2019;46(7):595–603. doi:10.1111/1346-8138.14933
39. Tan L, Zhao S, Zhu W, et al. The Akkermansia muciniphila is a gut microbiota signature in psoriasis. Exp Dermatol. 2018;27(2):144–149. doi:10.1111/exd.13463
40. Myers B, Brownstone N, Reddy V, et al. The gut microbiome in psoriasis and psoriatic arthritis. Best Pract Res. 2019;33(6):101494. doi:10.1016/j.berh.2020.101494
41. Bisanz JE, Upadhyay V, Turnbaugh JA, et al. Meta-analysis reveals reproducible gut microbiome alterations in response to a high-fat diet. Cell Host Microbe. 2019;26(2):265–272.e4. doi:10.1016/j.chom.2019.06.013
42. Jo J-K, Seo S-H, Park S-E, et al. Gut microbiome and metabolome profiles associated with high-fat diet in mice. Metabolites. 2021;11(8):482. doi:10.3390/metabo11080482
43. Sun M, Wu W, Liu Z, et al. Microbiota metabolite short chain fatty acids, GPCR, and inflammatory bowel diseases. J Gastroenterol. 2017;52(1):1–8. doi:10.1007/s00535-016-1242-9
44. Fang D, Shi D, Lv L, et al. Bifidobacterium pseudocatenulatum LI09 and Bifidobacterium catenulatum LI10 attenuate D-galactosamine-induced liver injury by modifying the gut microbiota. Sci Rep. 2017;7(1). doi:10.1038/s41598-017-09395-8
45. Mukherjee A, Lordan C, Ross RP, et al. Gut microbes from the phylogenetically diverse genus Eubacterium and their various contributions to gut health. Gut Microbes. 2020;12(1):1802866. doi:10.1080/19490976.2020.1802866
46. Zhou J, Li M, Chen Q, et al. Programmable probiotics modulate inflammation and gut microbiota for inflammatory bowel disease treatment after effective oral delivery. Nat Commun. 2022;13(1):1.
47. Li S, Zhou L, Zhang Q, et al. Genistein improves glucose metabolism and promotes adipose tissue browning through modulating gut microbiota in mice. Food Funct. 2022;13(22):11715–11732. doi:10.1039/D2FO01973F
48. Ma E, Maskarinec G, Lim U, et al. Long-term association between diet quality and characteristics of the gut microbiome in the multiethnic cohort study. Br J Nutr. 2022;128(1):93–102. doi:10.1017/S0007114521002968
49. Bui TPN, A SS, Lagkouvardos I, et al. Comparative genomics and physiology of the butyrate‐producing bacterium intestinimonas butyriciproducens. Environ Microbiol Rep. 2016;8(6):1024–1037. doi:10.1111/1758-2229.12483
50. Thomann AK, Wüstenberg T, Wirbel J, et al. Depression and fatigue in active IBD from a microbiome perspective—a Bayesian approach to faecal metagenomics. BMC Med. 2022;20(1). doi:10.1186/s12916-022-02550-7
51. Yue S, Zhao D, Peng C, et al. Effects of theabrownin on serum metabolites and gut microbiome in rats with a high-sugar diet. Food Function. 2019;10(11):7063–7080. doi:10.1039/C9FO01334B
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.