Back to Journals » Infection and Drug Resistance » Volume 14
Genomic and Transcriptomic Analysis of Colistin-Susceptible and Colistin-Resistant Isolates Identify Two-Component System EvgS/EvgA Associated with Colistin Resistance in Escherichia coli
Authors Wan F, Xu L, Ruan Z , Luo Q
Received 22 April 2021
Accepted for publication 15 June 2021
Published 28 June 2021 Volume 2021:14 Pages 2437—2447
DOI https://doi.org/10.2147/IDR.S316963
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Fen Wan,1 Linna Xu,1 Zhi Ruan,2 Qixia Luo3
1School of Laboratory Medicine, Hangzhou Medical College, Hangzhou, Zhejiang, People’s Republic of China; 2Department of Clinical Laboratory, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, 310016, People’s Republic of China; 3State Key Laboratory for Diagnosis and Treatment of Infectious Diseases; Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital of Medical School, College of medicine, Zhejiang University, Hangzhou, 310003, People’s Republic of China
Correspondence: Qixia Luo
State Key Laboratory for Diagnosis and Treatment of Infectious Diseases; Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital of Medical School, College of medicine, Zhejiang University, 79 Qingchun Road, Hangzhou, 310003, People’s Republic of China
Email [email protected]
Zhi Ruan
Department of Clinical Laboratory, Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, 3 East Qingchun Road, Hangzhou, 310016, People’s Republic of China
Email [email protected]
Purpose: Colistin is one of the last-resort antimicrobial agents that combat the increasing threat of multi-drug resistant (MDR) gram-negative bacteria. Based on the known mechanism of colistin resistance which contributes to chromosomal mutations involved in the synthesis and modification of lipopolysaccharide (LPS), we explored the regulatory genes mediate colistin resistance, by whole genome sequencing and transcriptome analysis.
Materials and Methods: In this study, a colistin-resistant (Colr) strain Escherichia coli ATCC 25922-R was generated from colistin-sensible (Cols) strain E. coli ATCC 25922 by colistin induction. We compared the genome and transcriptome sequencing result from Cols and Colr strain. MALDI-TOF mass spectrometry was used to detect LPS.
Results: Genomic analysis and complementation experiment demonstrated the PmrB amino acid substitution in ATCC 25922-R (L14R) conferred the colistin resistance phenotype. Results of RNA sequencing (RNA-Seq) and comparative transcriptome analysis indicated that the two-component system EvgS/EvgA is highly involved in the global regulation of colistin resistance.
Conclusion: This study demonstrated that PmrB L14R amino acid substitution resulted in colistin resistance, and two-component system EvgS/EvgA might participate in colistin resistance in E. coli.
Keywords: colistin resistance, Escherichia coli, PmrAB, EvgS/EvgA
Introduction
Multi-drug resistant (MDR) bacteria are a global healthcare concern responsible for increased morbidity and mortality, hospitalization, and huge financial loss. In order to combat increasing rise of MDR bacteria, various therapies have been developed to inhibit MDR bacterial infection including traditional antibiotics, phage therapies, monoclonal antibodies (mAbs), immune stimulation, vaccines and antimicrobial peptides.1–4 But there are very few phages available to doctors and the number of currently licensed mAbs is limited in clinical practice. In terms of traditional antibiotics, only a handful of last-line antibiotics, such as colistin and tigecycline, are proven to be effective in MDR bacteria treatment.5 Colistin (polymyxin E) is a decades-old antimicrobial agent, intensively used in veterinary medicine and animal husbandry.6 In humans, the global spread of MDR Gram-negative bacteria with limited therapeutic options has resulted in the return of colistin use regardless of its documented nephrotoxicity.7
Colistin is a cationic polypeptide that disrupts membrane integrity through displacement of cations like Mg2+ and Ca2+ in the outer membrane, leading to cell lysis.5,8 Two widely accepted mechanisms of colistin resistance that have been described in Gram-negative bacteria are as follows: chromosomal mutations in genes involved in the synthesis and modification of lipopolysaccharide (LPS) and the acquisition of mobile colistin resistance (mcr) genes encoding phosphoethanolamine transferases, resulting in a more cationic charged LPS or loss of LPS that confers reduced susceptibility to colistin.8
The critical step of colistin resistance is the modification of negatively charged lipid A, the endotoxic component, via diverse routes, particularly through derivation of lipid A phosphate moieties with a sugar or ethanolamine.9 The two-component regulatory systems PmrAB and PhoPQ regulate expression of the genes (pmrC, arn and eptA) that mediate phosphoethanolamine modification of lipid A, which results in a decreased binding to the bacterial outer membrane surface.10,11 Mutations in pmrAB, phoPQ have been identified in several colistin resistant bacteria, including Escherichia coli, Klebsiella pneumoniae and Acinetobacter baumannii.12–14 In the transcriptomes of colistin-resistant K. pneumoniae, an elevated expression of pmrH orperon was responsible for the biosynthesis of Ara4N and modification of the lipid A component.12 In A. baumannii, up-regulated expression of pmrAB and lpxB (enzyme involved in the lipid A biosynthesis pathway) genes associated with colistin resistance has also been reported.15
Recent studies of colistin resistance mostly focus on the plasmid-encoded mobilized colistin resistant gene mcr-1 in many aspects, such as its prevalence, antibiotic resistance mechanism, and its transmission between different bacteria. But in fact, chromosomal gene mutation mediated colistin resistance always possesses higher level of resistance and is not easily lost in the process of passage. Therefore, it is worth studying the mechanisms of chromosome mediated colistin resistance, especially the less studied regulation profile.
Here, by treating E. coli with consistent colistin stimulation which imitates the clinical treatment condition, we obtained the colistin-resistant strain E. coli ATCC 25922-R. Using genomic, transcriptomic, bioinformatics and other approaches to study the mechanism of chromosome-related colistin resistant genes, we successfully explored the genetic basis and analyzed the global transcription profile to figure out new genes responsible for colistin resistance. Our results revealed that it is highly possible that two-component system EvgS/EvgA participates in the regulation of colistin resistance in E. coli.
Materials and Methods
Strains and Antimicrobial Susceptibility Testing
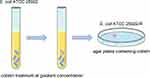
The strain E. coli ATCC 25922 was used to conduct this study. Laboratory evolution of mutant with reduced colistin susceptibility, E. coli ATCC 25922-R was generated by broth step-wise induction.16 Briefly, a single colony of E. coli ATCC 25922 was grown in Muller-Hinton (MH) broth overnight at 37°C, and cultures were diluted 1:100 in MH broth containing serially increasing concentrations of colistin, starting at the one-half MIC of the respective isolate, and doubling every 24 hours until bacterial growth was completely inhibited. Cultures were plated on MH agar plates and a colony, namely E. coli ATCC 25922-R was stored for the following study. Colistin MIC determination was performed by broth microdilution methods in accordance with the Clinical and Laboratory Standards Institute 2019 (CLSI 2019).
Genomic Analysis and Gene Complementation
Genomic DNA of E. coli ATCC 25922 and Colr strain E. coli ATCC 25922-R were extracted using Gentra Puregene Yeast/Bact. Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions, from overnight cultures. After library preparation, genome of ATCC 25922 was sequenced using the PacBio RS II and Illumina Hiseq platform (Illumina, San Diego, CA, USA), and the genome of ATCC 25922-R was sequenced by Illumina Hiseq platform. Library construction and sequencing was performed at Beijing Novogene Bioinformatics Technology Co. Ltd. The filtered reads were assembled to generate one contig without gaps by SMRT 2.3.0. The Illumina-generated contigs were mapped to the PacBio-generated contigs for correction. Coding genes were annotated using Rapid Annotation using Subsystem Technology (RAST). For the genome comparison, assembled genomes were annotated using Prokka Software for unified comparison.17 Then Roary, a high speed pan-genome pipeline was used to cluster the genes encoding complete protein sequences.18 Complementation of pmrB was conducted by plasmid pHGE-Ptac.19 The gene of interest was amplified and inserted into MCS of pHGE-Ptac under the control of the tac promoter. The resulting vectors were transferred into its corresponding mutant strain via electro-transformation.
Real-Time PCR
Overnight cultures of the strains were diluted 1:100 and subcultured in MH medium for ~3 hrs at 37°C (absorbance at 600 nm [OD600] ~ 0.6) with or without colistin. Cells were collected at 4°C by centrifuging at 10000 rpm for 1 min, and total RNA of E. coli ATCC 25922 and E. coli ATCC 25922-R were extracted with RNeasy Mini Kit (QIAGEN, Alameda, USA) as the production protocol told. RNA degradation and contamination were monitored on 1% agarose gels. cDNA was created from each RNA sample. RT-qPCR was performed using an ABI 7300 96-well RT-qPCR system (Applied Biosystems) with SYBR Premix Ex Taq II (cat. no. RR820A; TaKaRa). mRNA expression levels of genes pmrB, relA, evgS and evgA were examined using real-time PCR primers (EC-gene-RT-F/R) listed in Supplementary Table S1. Relative expression values of each gene were normalized against the 16S rRNA gene of E. coli ATCC 25922 using the standard curve method. All samples were tested in triplicate, and the data were represented as means±SD (standard deviation).
RNA-Seq Experiments
For mRNA sequencing, overnight cultures of the strains were diluted 1:100 and subcultured in MH medium to OD600 ~ 0.5. Then the culture was induced with (ATCC 25922-R) or without (ATCC 25922 and ATCC 25922-R) 2 mg/L colistin for half an hour. Total RNA was extracted with RNeasy Mini Kit (QIAGEN, Alameda, USA) as the production protocol told. cDNA libraries were constructed and were sequenced on an Illumina HiSeq platform. Reads from each strain were mapped to the corresponding genome assembly and RPKM (reads per kilobase of transcript per million mapped reads) values were calculated based on the length of the gene reads count mapped to the gene. The cutoff for gene expression was set at an RPKM value of at least 10. The raw sequencing data were deposited in the NCBI Sequence Read Archive (BioProject accession number PRJNA681352). Genes with significant expression RPKM values were identified. Gene transcripts with ≥ 2-fold change in expression and a q value of ≤ 0.05 were considered as differentially expressed. Gene ontology (GO) was analyzed using the GOseq R package. GO terms with p-value less than 0.5 were considered significantly enriched by differentially expressed genes. KEGG pathway mapping analysis was performed to examine the associated pathways and functions in the differentially expressed genes.
Extraction of LPS-Lipid A and MALDI-TOF Analysis
The crude LPS was isolated by hot phenol/water extraction.20 Briefly, overnight bacterial cultures were collected for LPS isolation. Bacterial cells (1~10 mg) were washed with Tris-HCl (30mM, pH 8.0) twice, centrifuged at 6000 rpm for 10 min, and resuspended in 0.4 mL of Tris-HCl containing 20% sucrose. Bacterial samples were incubated for 0.5 h after adding 40 μL of lysozyme (1mg/mL, 100 mM EDTA, pH 7.3), and frozen at −80°C for 0.5 h, then thawed at room temperature. Following several freezing and thawing cycles, the samples were lysed by sonication and cell lysate was removed for 15 min at 6000 rpm. The supernatant was further centrifuged at 16,000 rpm for 1 h to precipitate the crude LPS.
Then the crude LPS was freeze-dried and resuspended in a solution of 20 mM Tris-HCl with 0.2% SDS. DNase I (25 μg/mL) and RNase A (100 μg/mL) were added and the solution was incubated for 2h at 37°C after following a removal of protein contaminants via treatment with proteinase K at 37°C for 1h. To cleave the Kdo linkage, the crude LPS was heated in 10 mM sodium acetate buffer (pH 4.5) with aqueous 0.2% SDS at 100°C for 1 h and then precipitated with acidified ethanol (100 μL) to remove SDS by centrifugation at 5000 rpm for 5 min. The resulting pellets were washed with 95% ethanol twice and freeze-dried for storage.
Negative ion MALDI-TOF mass spectra were used to analyze the chemical structures of lipid A from different strains. Lipid A fractions were dissolved in 20 μL of chloroform/methanol (2:1) solution and mixed with 2,5-dihydroxybenzoic acid matrix in the solution of chloroform/methanol/water (3:1.5:0.25) (20 mg/mL). One microliter of lipid A solution was loaded to produce the MS spectrum. An average of 500 shots was summed into a single mass spectrum.
Results
Generation of Colistin Resistant Strain and Complementation
E. coli ATCC 25922 was treated with colistin to obtain colistin resistant strain E. coli ATCC 25922-R as described in materials and methods. Its flow chart was shown in Figure 1. The colistin MICs of E. coli ATCC 25922 and E. coli ATCC 25922-R were 0.25 mg/L and 64 mg/L, respectively. Comparative genomic analysis observed a mutation of PmrB (L14R) in ATCC 25922-R, which may responsible for the covalent modification of phosphate groups on lipid A. We also found an additional mutation in RelA (R137W), the GTP pyrophosphokinase, which is involved in the synthesis of guanosine 3ʹ,5ʹ-bispyrophosphate (ppGpp) during the stringent response to amino acid starvation21,22 (Figure 2A). Complementation of pmrB gene amplified from ATCC 25922 to ATCC 25922-R restored the susceptibility of ATCC 25922-R (colistin MIC of the complementary strain is 0.5 mg/L), which demonstrated that the point mutation of PmrB (L14R) in this study contributed to the colistin resistant phenotype of ATCC 25922-R. Therefore, the identification of the mutation in pmrB is involved in colistin resistance in ATCC 25922-R isolated after in vitro colistin exposure.
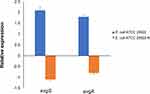
Expression Analysis of pmrB and relA
Based on the prior knowledge of pmrB function reported previously, we inferred that this mutation might cause the transcriptional alteration in the pmrAB system. To evaluate the effect of this mutation on the induced Colr strain, quantitative RT-PCR was performed using the primers designed for pmrB transcript. The Colr strain displayed 2.3-fold increased expression of pmrB, which is consistent with activating mutations in pmrB (Figure 2B). RT-PCR result showed no significant change in the expression of another mutated gene, relA (Figure 2B). Since lipid A modification has been associated with colistin resistance, we analyzed lipid A moieties from E. coli ATCC 25922-R by MALDI-TOF mass spectrometry. Peak at m/z 1914.5 corresponded to the addition of one phosphoethanolamine (pEtN) moiety and to the phosphate group of the native lipid A, leading to an increase of +123 mass units compared to the mass of the major peak of native lipid A at m/z 1797.4. Two additional peaks at m/z 1934.4 and m/z 2164.5 corresponded to the addition of 4-amino-L-arabinose (L-Ara4N) to the phosphate of the native lipid A, generating an increase of +131 mass units compared to the native lipid A (Figure S1). Both pEtN and L-Ara4N additions were detected in E. coli ATCC 25922-R, suggesting that pmrB mutation altered the lipid A structure with various forms.
Transcriptome Characterization
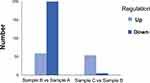
In order to gain a deeper insight into the mechanism of the E. coli ATCC 25922-R resistant to colistin and gene expression change under the clinical treatment of colistin, we performed RNA-seq analysis of three samples, E. coli ATCC 25922 (sample A), ATCC 25922-R (sample B), and ATCC 25922-R with colistin treatment under the dosage of 2 mg/L (sample C), a concentration that is used in clinical therapy with the aim to simulate medication in clinical practice, as described in materials and methods. Over 99% of all clean reads were aligned to reference genome of E. coli ATCC 25922. Each biological replicate was highly similar to each other (R2>0.95), which indicates the RNA-seq data were suitable for transcriptome analysis. The transcriptome comparisons were conducted between sample A and B, sample B and C. The expression profile in the two comparisons was shown in Figure 3. Hierarchical clustering in the differentially expressed genes was analyzed and shown in the format of heat map observed in Figure 4.
In the comparison of sample B vs sample A, there were 278 differentially expressed genes, of which 59 were up-regulated and 219 were down-regulated (Supplementary Table S2). Kyoto encyclopedia of genes and genomes (KEGG) was used to compare the transcriptome data. Most differentially expressed genes were focused on citrate cycle (TCA cycle) and oxidative phosphorylation, which indicates that these biological processes might be affected by colistin resistance (Figure 4A). The top ten up-regulated and ten most down-regulated genes are listed in Table 1. Among the most up-regulated genes, gene yibD (D1792_15900) encoding a glycosyltransferase responsible for the synthesis of LPS cores was shown to be significantly up-regulated in colistin resistant E. coli ATCC 25922-R. Elevated transcription of arn operon and eptA associated with lipopolysaccharide modification were observed as well, which might partly explain the enhanced colistin resistance trait in the induced Colr strain. Other components of the PmrAB regulon, such as pmrD, showed no significant change in both strains. Most of the highly down-regulated genes were involved in glycolate utilization, an important energy source which provides an energetic advantage to bacterial species. The glc operon, consisting of GlcB encoding malate synthase G and the gene encoding glycolate oxidase, exhibited notable decline in transcriptional levels. Other genes participating in TCA cycle, such as D1792_24720 encoding succinate dehydrogenase, were down-regulated obviously. From these results, we inferred that E. coli ATCC 25922-R may reduce unnecessary energy demands to maintain the colistin resistance phenotype.
 |
Table 1 Top Ten Up- and Down-Regulated E. coli ATCC 25922-R (Sample B) Genes in Comparison with E. coli ATCC 25922 (Sample A) |
In E. coli ATCC 25922-R and its counterpart treated with 2 mg/L colistin (sample C vs sample B), a distinct transcriptional pattern was observed. Only 54 up-regulated genes and 5 noticeably down-regulated genes were observed, characterized by the elevated transcription of glycosyltransferase, which is involved in colistin resistance (Supplementary Table S3). Top ten genes that exhibited significant expression levels are listed in Table 2. The Toll-like receptor homologous protein TcpC (D1792_06120) was greatly up-regulated in E. coli ATCC 25922-R grown with 2 mg/L colistin (Table 2). Combined with the fact that TcpC was identified as a virulence factor of E.coli and interfered with the innate immune response of the host by interrupting the NF-κB signaling pathway,23 we speculated that the pathogenicity of E. coli ATCC 25922-R might be enhanced when treated with colistin. Furthermore, the transcriptional levels of proteins like XcbB (D1792_12290) belonging to K-antigen gene cluster and PapX (D1792_11760) encoding a fimbrial-specific inhibitor were markedly increased, indicating that serological phenotype and motility might be affected (Table S2). The biological functions associated with differentially expressed genes were also analyzed in KEGG pathway (Figure 5B). Among them, “ubiquitin mediated proteolysis” was the most highly represented category.
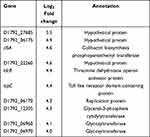 |
Table 2 Top Ten Up-Regulated Genes in E. coli ATCC 25922-R Resistant Treated with 2mg/L Colistin (Sample C) |
Based on the knowledge that membrane transporters and two component systems (TCS) are closely connected with colistin resistance, we searched for these genes that changed expression levels both in sample B vs A and sample C vs B groups (Table 3). Because there were only 59 genes that showed remarkably up-regulated expression, we set the value of Log2 fold change above 0.5 for screening condition in order to get a more comprehensive result about genes with changed expression level. The most distinctive difference was the expression of evgS (1.34-fold down-regulated and 0.71-fold up-regulated, respectively), encoding sensor histidine kinase EvgS of TCS EvgS/EvgA which confers acid resistance in E. coli. Moreover, we also noticed that the expression level of another AraC-XylS family transcription factor ydeO altered both in two groups, which was consistent with the previous research findings that ydeO expression is activated by TCS EvgS/EvgA.24 qRT-PCR experiment was performed to verify the transcriptome result (Figure 6). According to the report that activated EvgS/EvgA promoted expression of PhoP-activated genes,25,26 our data suggest that EvgS/EvgA might be involved in the colistin resistance by interacting indirectly with PhoQ/PhoP system.
 |
Table 3 Differentially Expressed Genes Both in Sample B vs A and Sample C vs B |
Discussion
Antibiotics like colistin are the last resort to treat clinical infections caused by carbapenem-resistant Enterobacteriaceae (CRE). However, the new emergence of resistance to colistin severely restricts therapeutic options. Among Enterobacteriaceae,E. coli tops the list in causing infections because of high prevalence, multidrug resistance and rapid transfer or acquisition of antimicrobial resistant traits through horizontal gene transfer. The mechanism of colistin resistance employed by the majority of bacteria can be explained in two ways: chromosomal point mutations in pmrA/pmrB and phoP/phoQ; and plasmid mediated colistin resistant mcr-1 and mcr-2 genes. Hence, most of the studies are focused on alterations in the expression and regulation mode of these genes. There are relatively few studies which have investigated colistin resistance in global transcriptome of E. coli. In this study, we aimed to identify two distinct transcription profiles by comparing whole genomic and transcriptomes from colistin-susceptible and colistin-resistant strains. The mutation at the position of 14th amino acid substitution of PmrB from Leu to Arg has been reported in E. coli previously,27,28 and this alteration might lead to a constitutive activation of PmrB sensor that is responsible for colistin resistance in E. coli, which implicates the ability of clinical strains to evolve to a Colr phenotype. The mechanism by which the Leu14Arg replacement alters the functions of PmrB remains unknown. This mutation is located at the amino-terminal protein portion of cytoplasmic secretion signal domain of PmrB, which remarkably altered the structure of the cytoplasmic domain and downstream regions. The mutations in the PmrB have previously been reported to be associated with acquisition of a colistin resistance phenotype in other pathogenic species including P. aeruginosa, A. baumannii and E. coli, underscoring the role of PmrB as an important mutational target in the evolution of colistin resistance in Gram-negative pathogens.14,29,30 In addition, our previous data indicated that mutations in PmrB would alter the expression level of pmrAB operon and its downstream genes in the mutants.18
Previous studies have revealed that Pmr mutation leads to the composition alteration of lipid A in several gram-negative bacteria, which reduces LPS affinity for colistin by reducing positive charges.8,31 As we expected, our data also showed the lipid A modification with L-Ara4N and pEtN in Colr strains, which indicates that lipid A aminoarabinosylation would be sufficient to confer colistin resistance.
The comparative transcriptome analysis discovered interesting genes that might be involved in colistin resistance in E. coli. Transcriptional level of gene yibD encoding glycoslytransferases regulated by PmrA increased nearly 16-fold in Colr strain, which implicates increased production of LPS synthesized by the sequential addition of sugar moieties by glycosyltransferases. It is worth noting that the expression of two-component system EvgS/EvgA was strongly affected in E. coli ATCC 25922-R. The EvgS/EvgA two-component system signal transduction regulates genes encoding the glutamate-dependent acid resistance system and a number of drug efflux pumps in response to low pH and antibiotic stress.
Several studies have revealed that EvgS/EvgA system interacts with PhoQ/PhoP system by enhancing the expression of PhoP regulons via the small connecter protein SafA, signal transduction cascade between EvgS/EvgA and PhoQ/PhoP TCSs in E. coli.25 Another report showed that overexpression of EvgA is known to induce multidrug resistance by up-regulating the multi-drug efflux pump genes.32 Moreover, a study reported that regulator YdeO activated by EvgS also contributes to multi-drug resistance through the activation of MdtEF efflux pump.33 Therefore, combined with these studies, we hypothesize that EvgS/EvgA might participate in colistin resistance either by regulating the multi-drug efflux protein or by interacting with TCS PhoQ/PhoP in E. coli. The biological function of the EvgS/EvgA system in colistin resistance needs further investigation. These findings will contribute to our understanding of the mechanism underlying the occurrence of colistin resistance.
Conclusion
In conclusion, our findings revealed two new mutations in the genome of colistin-resistant E. coli, and analyzing transcriptome profiles of colistin-resistant E. coli under the treatment of colistin, which indicated that two-component system EvgS/EvgA might be responsible for colistin resistance in E. coli.
Acknowledgment
We thank Hao Fu, for the technical support and quality control.
Funding
This research was funded by the National Key R&D Program of China(2017YFC1600100), National Natural Science Foundation of China (81702040), the National Science Foundation of Zhejiang province, China (LY20H190002) to L.Q., the Scientific Research Foundation of Hangzhou Medical College (Grant No. KYYB202004) to F.W.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Biswas S, Brunel JM, Dubus JC, Reynaud-Gaubert M, Rolain JM. Colistin: an update on the antibiotic of the 21st century. Expert Rev Anti Infect Ther. 2012;10:917–934.
2. Brives C, Pourraz J. Phage therapy as a potential solution in the fight against AMR: obstacles and possible futures. Palgrave Commun. 2020;6:100.
3. Pelfrene E, Mura M, Cavaleiro Sanches A, Cavaleri M. Monoclonal antibodies as anti-infective products: a promising future? Clin Microbiol Infect. 2019;25:60–64.
4. Czaplewski L, Bax R, Clokie M, et al. Alternatives to antibiotics-a pipeline portfolio review. Lancet Infect Dis. 2016;16:239–251.
5. Bialvaei AZ, Samadi Kafil H. Colistin, mechanisms and prevalence of resistance. Curr Med Res Opin. 2015;31:707–721.
6. Li J, Nation RL, Turnidge JD, et al. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect Dis. 2006;6:589–601.
7. Tran TB, Velkov T, Nation RL, et al. Pharmacokinetics/pharmacodynamics of colistin and polymyxin B: are we there yet?. Int J Antimicrob Agents. 2016;48:592–597.
8. Olaitan AO, Morand S, Rolain JM. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol. 2014;5:643.
9. Raetz CR, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in gram-negative bacteria. Annu Rev Biochem. 2007;76:295–329.
10. Gunn JS. The Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol. 2008;16:284–290.
11. Quesada A, Porrero MC, Téllez S, Palomo G, García M, Domínguez L. Polymorphism of genes encoding PmrAB in colistin-resistant strains of Escherichia coli and Salmonella enterica isolated from poultry and swine. J Antimicrob Chemother. 2015;70:71–74.
12. Wright MS, Suzuki Y, Jones MB, et al. Genomic and transcriptomic analyses of colistin-resistant clinical isolates of Klebsiella pneumoniae reveal multiple pathways of resistance. Antimicrob Agents Chemother. 2015;59:536–543.
13. Park YK, Lee JY, Ko KS. Transcriptomic analysis of colistin-susceptible and colistin-resistant isolates identifies genes associated with colistin resistance in Acinetobacter baumannii. Clin Microbiol Infect. 2015;21:
14. Adams MD, Nickel GC, Bajaksouzian S, et al. Resistance to colistin in Acinetobacter baumannii associated with mutations in the PmrAB two-component system. Antimicrob Agents Chemother. 2009;53:3628–3634.
15. Martínez-Guitián M, Vázquez-Ucha JC, Álvarez-fraga L, et al. Antisense inhibition of lpxB gene expression in Acinetobacter baumannii by peptide-PNA conjugates and synergy with colistin. J Antimicrob Chemother. 2020;75:51–59.
16. Luo Q, Niu T, Wang Y, et al. In vitro reduction of colistin susceptibility and comparative genomics reveals multiple differences between MCR-positive and MCR-negative colistin-resistant Escherichia coli. Infect Drug Resist. 2019;12:1665–1674.
17. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069.
18. Page AJ, Cummins CA, Hunt M, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693.
19. Luo Q, Dong Y, Chen H, Gao H. Mislocalization of Rieske protein PetA predominantly accounts for the aerobic growth defect of Tat mutants in Shewanella oneidensis. PLoS One. 2013;8:e62064.
20. Caroff M, Tacken A, Szabo´ L. Detergent-accelerated hydrolysis of bacterial endotoxins and determination of the anomeric configuration of the glycosyl phosphate present in the “isolated lipid A” fragment of the Bordetella pertussis endotoxin. Carbohydr. Res. 1988;175:273–282.
21. Hogg T, Mechold U, Malke H, Cashel M, Hilgenfeld R. Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response. Cell. 2004;117:57–68.
22. Viducic D, Ono T, Murakami K, et al. Functional analysis of spoT, relA and dksA genes on quinolone tolerance in Pseudomonas aeruginosa under nongrowing condition. Microbiol Immunol. 2006;50:349–357.
23. Schubert S, Nörenberg D, Clermont O, et al. Prevalence and phylogenetic history of the TcpC virulence determinant in Escherichia coli. Int J Med Microbiol. 2010;300:429–434.
24. Eguchi Y, Utsumi R. Alkali metals in addition to acidic pH activate the EvgS histidine kinase sensor in Escherichia coli. J Bacteriol. 2014;196:3140–3149.
25. Eguchi Y, Okada T, Minagawa S, et al. Signal transduction cascade between EvgA/EvgS and PhoP/PhoQ two-component systems of Escherichia coli. J Bacteriol. 2004;186:3006–3014.
26. Eguchi Y, Ishii E, Hata K, et al. Regulation of acid resistance by connectors of two-component signal transduction systems in Escherichia coli. J Bacteriol. 2011;193:1222–1228.
27. Choi Y, Lee JY, Lee H, et al. Comparison of Fitness Cost and Virulence in Chromosome- and Plasmid-Mediated Colistin-Resistant Escherichia coli. Front Microbiol. 2020;13(11):798.
28. Liao W, Lin J, Jia H, et al. Heteroresistance to Colistin in Escherichia coli Isolates from Wenzhou, China. Infect Drug Resist. 2020;9(13):3551–3561.
29. Abraham N, Kwon DH. A single amino acid substitution in PmrB is associated with polymyxin B resistance in clinical isolate of Pseudomonas aeruginosa. FEMS Microbiol Lett. 2009;298:249–254.
30. Sato T, Shiraishi T, Hiyama Y, et al. Contribution of novel amino acid alterations in PmrA or PmrB to colistin resistance in mcr-negative Escherichia coli clinical isolates, including major multidrug-resistant lineages O25b: H4-ST131-H30Rxand Non-x. Antimicrob Agents Chemother. 2018;62:e00864–18.
31. Baron S, Hadjadj L, Rolain JM, Olaitan AO. Molecular mechanisms of polymyxin resistance: knowns and unknowns. Int J Antimicrob Agents. 2016;48:583–591.
32. Itou J, Eguchi Y, Utsumi R. Molecular mechanism of transcriptional cascade initiated by the EvgS/EvgA system in Escherichia coli K-12. Biosci Biotechnol Biochem. 2009;73:870–878.
33. Nishino K, Senda Y, Hayashi-Nishino M, Yamaguchi A. Role of the AraC-XylS family regulator YdeO in multi-drug resistance of Escherichia coli. J Antibiot (Tokyo). 2009;62:251–257.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.