Back to Journals » Cancer Management and Research » Volume 11
Genetic polymorphisms in IL-7 and IL-7R are correlated with lung cancer risk in the Chinese Han population
Authors Zhang C, Su P , Chen W, Li Q, Dai R, Cheng YJ, Yang J
Received 24 January 2019
Accepted for publication 30 April 2019
Published 11 June 2019 Volume 2019:11 Pages 5393—5401
DOI https://doi.org/10.2147/CMAR.S202839
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Rituraj Purohit
Chan Zhang,1 Pincan Su,2 Wanlu Chen,1 Qi Li,1 Run Dai,1 YuJing Cheng,1 Jiangcun Yang3
1Department of Blood Transfusion, The First People’s Hospital of Yunnan Province, Kunming, Yunnan 650032, People’s Republic of China; 2Laboratory of Blood Transfusion, Yunnan Kunming Blood Center, Kunming, Yunnan 650106, People’s Republic of China; 3Department of Transfusion Medicine, Shanxi Provincial People’s Hospital, Xi’an, Shaanxi 710068, People’s Republic of China
Purpose: IL-7/IL-7R axis participates in the initiation and progression of lung cancer (LC). This study aimed to explore the potential influence of IL-7/IL-7R polymorphisms on LC risk.
Patients and methods: In total, 1,010 participants (507 LC patients and 503 healthy controls) were enrolled. Five single-nucleotide polymorphisms (SNPs) in IL-7R and one SNP in IL-7 were genotyped in included samples with Agena MassARRAY system. OR and 95% CIs were computed by logistic regression analysis after adjusting for age and gender. Stratified analyses with demographic and clinical characteristics were also performed. Finally, linkage disequilibrium (LD) analysis was conducted with the PLINK version 1.07 software.
Results: IL-7R rs10053847 variant was related to a decreased LC risk under the allele gene (OR =0.78, P=0.043) and additive model (OR =0.77, P=0.042). The results of stratified analysis indicated that this SNP was associated with a lower LC risk among nonsmokers (AA/GG: OR =0.09, P=0.033; AA/AG+GG: OR =0.10 P=0.037) or nondrinkers (AA/GG: OR =0.07, P=0.047; AA/AG+GG: OR =0.18 P=0.049). Moreover, carriers of IL-7R rs10213865-C allele had an increased lung adenocarcinoma risk (CA/AA: OR =1.60, P=0.011; CC+CA/AA: OR =1.62, P=0.007; CA/CA/AA: OR =1.50, P=0.007). Additionally, AGAA haplotype (rs10213865, rs969129, rs118137916 and rs10053847) increased LC risk (OR =1.30, P=0.041).
Conclusion: IL-7R rs10053847 was correlated with a decreased LC risk, while IL-7R rs10213865 was correlated with an elevated lung adenocarcinoma risk, implying these two SNPs might play essential roles in LC risk evaluation.
Keywords: lung cancer, IL-7R, IL-7, polymorphisms, cancer susceptibility, case-control study
Introduction
Lung cancer (LC) has been regarded as one of the multifactorial disorders and remained a considerable public health challenge around the world.1 It is estimated that approximately 2.09 million cases were diagnosed worldwide in 2018.2 Moreover, the mortality rate is relatively higher in China than most other countries and predicted to continuously increase before 2030.3 Currently, although numerous investigators have emphasized that genetic and environmental factors participate in the progression of LC,4,5 the detailed pathogenesis of LC has not been fully elaborated. Several research groups claimed that there were strong associations between genetic variations and the occurrence and development of LC in the past few years.6,7
IL-7, a pleiotropic cytokine primarily secreted by stromal cells, is involved in the several cell biological processes such as lymphangiogenesis, cell proliferation and apoptosis through binding to its receptor (IL-7R).8,9 Increasing studies have found that IL-7 and IL-7R participate in the development of various cancers including LC by mediating multiple cell signaling pathways.10,11 Andersson et al previously found that IL-7 could significantly diminish tumor burdens via enhancing the specific chemokine receptor-dependent T-cell antitumor activity in LC progression.12 Jian et al pointed out that IL-7R interacted with some autophagy-associated molecules to activate the signaling pathways in cell apoptotic process of LC.13 Liu et al also argued that IL-7/IL-7R axis might be responsible for LC cell apoptosis regulation and survival outcomes.14 Recently, overwhelming evidence has explored the relationships between SNPs (single-nucleotide polymorphisms) in many tumor-related genes and LC risk.15,16 Van Dyke et al suggested that IL7R rs1494555 and rs7737000 variants were significantly linked to a higher risk of non-small LC among Caucasians.17 However, the underlying impact of IL-7/IL-7R polymorphisms (rs10213865, rs969129, rs118137916, rs10053847 and rs6451231 in IL-7R and rs117173992 in IL-7) on the prevalence of LC has not been clarified.
Therefore, we carried out an association analysis based on a Chinese Han population to evaluate the potential correlations between IL-7/IL-7R SNPs and the risk of LC.
Materials and methods
Study subjects
A total of 1,010 participants (507 patients suffering from LC and 503 cancer-free controls) were consecutively enrolled from Shaanxi Provincial Cancer Hospital. All cases were newly diagnosed and histologically confirmed lung carcinoma. Patients did not receive chemotherapy or radiotherapy before collecting samples. Patients with prior cancer history were excluded from this study. Meanwhile, all control subjects underwent medical examinations in the same hospital and did not have any family history of LC or other diseases. All eligible participants were all genetically unrelated to each other. The detailed demographic and the clinical data were subsequently obtained according to the pre-established standardized questionnaire, medical record and/or the face-to-face interviews, primarily including age, gender, body mass index (BMI), smoking and drinking status, pathological type, lymph nodes metastasis and TNM staging. This work was supported by Ethics Committee of Shaanxi Provincial Cancer Hospital and the written informed consent was acquired from each participant. All experiments were conducted in accordance with the World Medical Association Declaration of Helsinki.
SNP selection and genotyping
Two cancer-associated genes (IL-7R and IL-7) were selected to evaluate the correlation between their polymorphisms and the susceptibility to LC. Peripheral blood samples (5 mL) were collected from each subject, and genomic DNA was extracted using GoldMag whole-blood genomic DNA purifcation kit (GoldMag Co. Ltd., Xiʹan, China) according to the manufacturer′s recommendations. The candidate SNPs of IL-7R and IL-7 were selected based on the 1,000 Genomes Project database (
Statistical analyses
We utilized the Pearson′s χ2 test and Student’s t test to assess differences in demographic data (age, gender, BMI, smoking and drinking status) between cases and controls. Hardy–Weinberg equilibrium (HWE) analyses of each SNP were performed by comparing the observed and expected genotype frequencies among controls using the Fisher’s exact test. ORs and their 95% CIs were estimated by the logistic regression analysis with adjustment for age and gender. Multiple genetic models (dominant, recessive and additive model) were adopted to explore the relationships of IL-7R and IL-7 polymorphisms and LC risk using PLINK v1.07 software. Additionally, the stratified analyses in terms of several confounding factors such as age, gender, body mass index (BMI), smoking and drinking status, pathological type, lymph nodes metastasis and TNM stage were also conducted. Finally, the pairwise LD and haplotype analysis were carried out using PLINK v1.07 software and Haploview v4.2 software. All statistical analyses were performed by SPSS v 18.0 software (Armonk, New York City, NY, USA), and two-sided P<0.05 indicated statistical significance. We used power and sample size (PS) calculation software (
Results
Participant characteristics and SNP identification
As exhibited in Table 1, there were 507 LC patients (352 males and 155 females; median age: 60.79±9.96 years) and 503 healthy controls (354 males and 149 females; median age: 59.94±9.58 years) in the present work. No significantly statistical differences on age, gender and smoking status were observed between cases and control groups (age: P=0.164; gender: P=0.742; smoking status: P=0.164; Table 1). However, BMI and drinking status were dramatically different between the two groups (P<0.001). Five candidate SNPs in IL-7R (rs10213865, rs969129, rs118137916, rs10053847 and rs6451231) and one SNP in IL-7 (rs117173992) were successfully genotyped as displayed in Table 2. The observed genotype frequency of all tested SNPs in control groups was strongly accorded with HWE (P>0.05). Notably, the frequency distribution of IL-7R rs10053847-A allele was remarkably lower in LC patients than health controls (OR =0.78, 95% CI: 0.62–0.99, P=0.043, power =52.50%), suggesting that this SNP served as a protective factor for the susceptibility to LC.
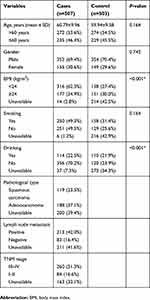 | Table 1 Characteristics of cases and cancer-free controls |
 | Table 2 Basic characteristics about candidate SNPs and associations with the risk of lung cancer in allele mode |
LC risk evaluation
To further evaluate the correlations of SNPs in IL-7R and IL-7 with LC risk, multiple inheritance models (genotype, dominant, recessive and additive models) were performed using logistic regression analyses with adjustment for age and gender (Table 3). The results also showed that carriers of IL-7R rs10053847 mutant allele had a decreased LC risk under additive model (OR =0.77, 95% CI: 0.60–0.99, P=0.042). No significant associations were detected between other SNPs and the susceptibility to LC (P>0.05).
 | Table 3 Relationships of polymorphisms in IL-7R and IL-7 genes and lung cancer susceptibility |
Stratified analyses were carried out to estimate relationships between these polymorphisms and several demographic characteristics (Tables 4 and
 | Table 4 The associations between IL-7R rs10053847 and the risk of lung cancer stratified by smoking and drinking status |
Additionally, carriers of IL-7R rs10053847 mutant allele had a lower risk of lung adenocarcinoma in additive model (OR =0.67, 95% CI: 0.47–0.95, P=0.025; Table 5). Conversely, IL-7R rs10213865 polymorphism was observed with a dramatically elevated the incidence of lung adenocarcinoma (CA/AA: OR =1.60, 95% CI: 1.11–2.29, P=0.011, power =93.15%; CC+CA/AA: OR =1.62, 95% CI: 1.14–2.29, P=0.007, power =95.33%; CC/CA/AA: OR =1.50, 95% CI: 1.11–2.01, P=0.007; Table 5) according to the stratification analysis with pathology type (Table 5 and
 | Table 5 Relationships of IL-7R rs10053847 and rs10213865 and pathology type risk of lung cancer |
Haplotype analysis of IL-7R polymorphisms
Linkage disequilibrium (LD) and corresponding haplotypes of IL-7R SNPs were further analyzed to estimate the association between this gene and the prevalence of LC. The results suggested that the high LD block was composed of four IL-7R polymorphisms (rs10213865, rs969129, rs118137916 and rs10053847) which formed five haplotypes (AGAA, ATGG, CGAG, AGAG and ATAG; Figure 1). Furthermore, logistic regression analysis based on haplotype was performed to investigate the impact of these haplotypes on the incidence of LC. We noted that there was a significant correlation between AGAA haplotype and an increased LC risk (OR =1.30, 95% CI: 1.01–1.66, P=0.041; Table 6).
 | Table 6 Associations of haplotype of IL-7R and the risk of lung cancer |
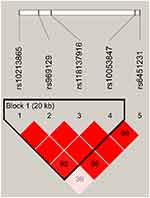 | Figure 1 The haplotype block map for single-nucleotide polymorphisms in the IL-7R. |
Discussion
In this present work, we first examined the relationships between IL-7/IL-7R polymorphisms and the risk of LC in a Chinese Han population. Our findings revealed that IL-7R rs10053847 and rs10213865 variants were predominately correlated with LC susceptibility, while no significant association of IL-7 polymorphism and LC risk was observed. To our best knowledge, this is the first study to provide evidence ofthe potential role of the IL-7R variants in LC risk. Combined with the previous studies,21 this association may be a promising starting point on the association of IL-7R polymorphism with LC formation and progression and provide data for the construction of a genetic panel for the prediction of LC risk in China.
IL-7R, located on chromosome 5p13, plays crucial roles in human diseases including malignancies. IL-7R has been demonstrated to be implicated with the molecular mechanisms of LC.22,23 Ming et al found that IL-7/IL-7R might promote lymphangiogenesis in LC by increasing the expression levels of specific vascular endothelial growth factor and stimulating the c-Fos/c-Jun pathway.24 Another research suggested that IL-7/IL-7R could elevate cyclin D1 expression through activating the AP1 signaling pathway to accelerate LC cell proliferation.25 More interestingly, numerous studies highlighted that IL-7R polymorphisms were involved in the initiation and development of several diseases in the recent years.26–28 However, the potential influence of IL-7R variants on the prevalence of LC has not been uncovered.
Herein, we conducted an association analysis between IL-7/IL-7R polymorphisms and LC risk in a Chinese Han population and found that IL-7R rs10053847 dramatically decreased the risk of LC under the additive model. rs10053847-A allele was predominately correlated with a lower incidence of LC among nonsmokers or nondrinkers, which further provided evidence that there was correlative effect between IL-7R polymorphisms and tobacco smoke exposure on the risk of LC.29 In addition, this SNP also significantly reduced the risk of lung adenocarcinoma in additive model. Therefore, we speculated that an allele of IL-7R rs10053847 acted as protective factors against LC occurrence. However, our stratified analysis revealed that another SNP in IL-7R (rs10213865) increased the susceptibility to lung adenocarcinoma under the heterozygous mutation, dominant and additive models. Moreover, there no significant difference was observed in other clinical features. Thus, it was inferred that IL-7R rs10213865 variant might only confer the risk for developing lung adenocarcinoma. More notably, the haplotype analysis implied that AGAA haplotype (rs10213865, rs969129, rs118137916 and rs10053847) was associated with a decreased LC risk, which further supported the conclusion that these two polymorphisms (rs10213865 and rs10053847) in IL-7R played vital roles in LC risk assessment. Although IL-7 was also found to participate in the pathogenesis of LC,30–32 there was no correlation between IL-7 rs117173992 and the risk of LC. Additional analyses of the relationship between this SNP and the incidence of LC are still required to verify in the future.
Certainly, there are still limitations in the current study. First, the study was performed using the hospital-based samples; therefore, the sample bias might be confounding factors for our findings. Second, the exhaustive functional analysis of these SNPs should be investigated to explain our results. Finally, the prognostic analysis of selected IL-7/IL-7R polymorphisms is needed to carry out in order to evaluate the survival outcomes of LC patients with these SNPs.
Conclusion
In conclusion, our results implied that IL-7R rs10053847 polymorphism was closely related to a decreased risk of LC, whereas carriers of IL-7R rs10213865-C allele showed an elevated adenocarcinoma risk, which would provide new evidence for prevention and diagnosis of LC. However, relevant functional study still needs to be undertaken in future.
Acknowledgments
We are grateful to the individuals who participated in this study. We also thank the clinicians and hospital staff who contributed to the sample and data collection for this study. We would also like to thank all those who contributed to this manuscript. This work was supported by the General Program of Applied Basic Research Projects-Joint Special Foundation from the Yunnan Provincial Science and Technology Department and the Kunming Medical University (2017FE468(−125)).
Disclosure
The authors report that they have no conflicts of interest in regard to this work.
References
1. Siegel RLMK, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
2. Cao M, Chen W. Epidemiology of lung cancer in China. Thorac Cancer. 2019;10(1):3–7. doi:10.1111/1759-7714.12916
3. Martín-Sánchez JCLN, González-Marrón A, Lidón-Moyano C, et al. Projections in breast and lung cancer mortality among women: a bayesian analysis of 52 countries worldwide. Cancer Res. 2018;78(15):4436–4442. doi:10.1158/0008-5472.CAN-18-0187
4. Gibelin CCS. Somatic alterations in lung cancer: do environmental factors matter? Lung Cancer. 2016;100:45–52. doi:10.1016/j.lungcan.2016.07.015
5. Gouvinhas CDMR, Oliveira D, Castro-Lopes JM, et al. Lung cancer: a brief review of epidemiology and screening. Future Oncol. 2018;14(6):567–575. doi:10.2217/fon-2017-0486
6. Clemenceau ABO, Landry-Truchon K, Lamontagne M, et al. Lung cancer susceptibility genetic variants modulate HOXB2 expression in the lung. Int J Dev Biol. 2018;62(11–12):857–864. doi:10.1387/ijdb.180210yb
7. Pintarelli G, Cotroneo CE, Noci S, et al. Genetic susceptibility variants for lung cancer: replication study and assessment as expression quantitative trait loci. Sci Rep. 2017;7:42185. doi:10.1038/srep42185
8. Wu LLJ, Xu HL, Xu B, Tong XH, Kwak-Kim J, Liu YS. IL-7/IL-7R signaling pathway might play a role in recurrent pregnancy losses by increasing inflammatory Th17 cells and decreasing Treg cells. Am J Reprod Immunol. 2016;76(6):454–464. doi:10.1111/aji.12588
9. Silva ALA, Martins LR, Cardoso BA, et al. IL-7 contributes to the progression of human T-cell acute lymphoblastic leukemias. Cancer Res. 2011;71(14):4780–4789. doi:10.1158/0008-5472.CAN-10-3606
10. Qu HZZ, Pan Z, Zhang T, Deng N, Chen G, Wang Z. IL-7/IL-7 receptor axis stimulates prostate cancer cell invasion and migration via AKT/NF-κB pathway. Int Immunopharmacol. 2016;40:203–210. doi:10.1016/j.intimp.2016.08.017
11. Andersson A1SM, Harris-White M, Huang M, et al. Role of CXCR3 ligands in IL-7/IL-7R alpha-Fc-mediated antitumor activity in lung cancer. Clin Cancer Res. 2011;17(11):3660–3672. doi:10.1158/1078-0432.CCR-10-3346
12. Andersson AYS, Huang M, Zhu L, et al. IL-7 promotes CXCR3 ligand-dependent T cell antitumor reactivity in lung cancer. J Immunol. 2009;182(11):6951–6958. doi:10.4049/jimmunol.0803340
13. Jian MYZ, Zhiying D, Yanduo J, Guocheng J. Interleukin 7 receptor activates PI3K/Akt/mTOR signaling pathway via downregulation of Beclin-1 in lung cancer. Mol Carcinog. 2018. doi:10.1002/mc.22933
14. Liu ZHWM, Ren HJ, Qu W, et al. Interleukin 7 signaling prevents apoptosis by regulating bcl-2 and bax via the p53 pathway in human non-small cell lung cancer cells. Int J Clin Exp Pathol. 2014;7(3):870–881.
15. Yan SSR, Wu S, Jin T, et al. Single nucleotide polymorphism in the 3ʹ untranslated region of LPP is a risk factor for lung cancer: a case-control study. BMC Cancer. 2019;19(1):35. doi:10.1186/s12885-018-5241-5
16. Nikseresht MSM, Dehghani M, Abidi H, et al. Association of single nucleotide autophagy-related protein 5 gene polymorphism rs2245214 with susceptibility to non-small cell lung cancer. J Cell Biochem. 2018. doi:10.1002/jcb.27467
17. Van Dyke AL, Cote ML, Wenzlaff AS, et al. Cytokine and cytokine receptor single-nucleotide polymorphisms predict risk for non-small cell lung cancer among women. Cancer Epidemiol Biomarkers Prev. 2009;18(6):1829–1840. doi:10.1158/1055-9965.EPI-08-0962
18. Dai ZJLX, Kang HF, Wang XJ, et al. Genetic variation in metastasis-associated in colon cancer-1 and the risk of breast cancer among the chinese han population: a strobe-compliant observational study. Medicine. 2016;95(6):e2801. doi:10.1097/MD.0000000000004864
19. Xia PLB, Geng T, Deng Z, et al. FGFR2 gene polymorphisms are associated with breast cancer risk in the Han Chinese population. Am J Cancer Res. 2015;5(5):1854–1861.
20. Dupont WD, Plummer WD
21. Cavic M, Spasic J, Krivokuca A, et al. TP53 and DNA-repair gene polymorphisms genotyping as a low-cost lung adenocarcinoma screening tool. J Clin Pathol. 2019;72(1):75–80. doi:10.1136/jclinpath-2018-205553
22. Suzuki KKK, Sima CS, Nitadori J, et al. Clinical impact of immune microenvironment in stage I lung adenocarcinoma: tumor interleukin-12 receptor β2 (IL-12Rβ2), IL-7R, and stromal FoxP3/CD3 ratio are independent predictors of recurrence. J Clin Oncol. 2013;31(4):490–498. doi:10.1200/JCO.2012.45.2052
23. Jian MQZ, Yanduo J, Guocheng J, Xueshan Q. Anti-lymphangiogenesis effects of a specific anti-interleukin 7 receptor antibody in lung cancer model in vivo. Mol Carcinog. 2015;54(2):148–155. doi:10.1002/mc.22082
24. Ming JZQ, Qiu X, Wang E. Interleukin 7/interleukin 7 receptor induce c-Fos/c-Jun-dependent vascular endothelial growth factor-D up-regulation: a mechanism of lymphangiogenesis in lung cancer. Eur J Cancer. 2009;45(5):866–873. doi:10.1016/j.ejca.2008.12.006
25. Ming JJG, Zhang Q, Qiu X, Wang E. Interleukin-7 up-regulates cyclin D1 via activator protein-1 to promote proliferation of cell in lung cancer. Cancer Immunol Immunother. 2012;61(1):79–88. doi:10.1007/s00262-011-1078-3
26. Kim JYCH, Kim HJ, Kim LH, Namgoong S, Shin HD. Association analysis of IL7R polymorphisms with inflammatory demyelinating diseases. Mol Med Rep. 2014;9(2):737–743. doi:10.3892/mmr.2013.1863
27. Heron MGJ, van Moorsel CH, Ruven HJ, Huizinga TW, van der Helm-van Mil AH, Claessen AM, van den Bosch JM. Variation in IL7R predisposes to sarcoid inflammation. Genes Immun. 2009;10(7):647. doi:10.1038/gene.2009.55
28. Safaei SPZ, Moin M, Houshmand M. IL7R and RAG1/2 genes mutations/polymorphisms in patients with SCID. Iran J Allergy Asthma Immunol. 2011;10(2):129–132. doi:10.02/ijaai.129132
29. Bao WLSH, Zhang AQ, Kong XM, Deng DH, Zhang YJ. Lack of associations of polymorphisms of IL-7R, IL-13 and IL-15 with NSCLCs in non-smoking Chinese. Asian Pac J Cancer Prev. 2011;12(12):3239–3244.
30. Wang YSC, Hough KP, Tousif S, et al. Myeloid-derived suppressor cells impair B cell responses in lung cancer through IL-7 and STAT5. J Immunol. 2018;201(1):278–295. doi:10.4049/jimmunol.1701069
31. Shiels MSPR, Hildesheim A, Engels EA, et al. Circulating inflammation markers and prospective risk for lung cancer. J Natl Cancer Inst. 2013;105(24):1871–1880. doi:10.1093/jnci/djt309
32. Suzuki TKH, Abe R. Requirement of interleukin 7 signaling for anti-tumor immune response under lymphopenic conditions in a murine lung carcinoma model. Cancer Immunol Immunother. 2016;65(3):341–354. doi:10.1007/s00262-016-1808-7
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
