Back to Journals » Infection and Drug Resistance » Volume 12
Genetic characterization of two vancomycin-resistant Staphylococcus aureus isolates in Kerman, Iran
Authors Ziasistani M , Shakibaie MR , Kalantar-Neyestanaki D
Received 16 February 2019
Accepted for publication 29 May 2019
Published 4 July 2019 Volume 2019:12 Pages 1869—1875
DOI https://doi.org/10.2147/IDR.S205596
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sahil Khanna
Mahsa Ziasistani1,2, Mohammad Reza Shakibaie3,4, Davood Kalantar-Neyestanaki3
1Student Research Committee, Faculty of Medicine, Kerman University of Medical Sciences, Kerman, Iran; 2Pathology and Stem Cell Research Center٫ Faculty of Medicine, Kerman University of Medical Sciences, Kerman, Iran; 3Department of Microbiology and Virology, Faculty of Medicine, Kerman University of Medical Sciences, Kerman, Iran; 4Research Center for Infectious Diseases and Tropical Medicine, Kerman University of Medical Sciences, Kerman, Iran
Aim: The aim of this study was the genetic characterization of two clinical vancomycin-resistant Staphylococcus aureus (VRSA) isolates.
Materials and methods: Resistance to vancomycin was determined by phenotypic method. PCR was used for detection of mecA, vanA, ermA, ermB, ermC, msrA/B, aph(2”)-Ic, aph(3ʹ)-IIIa, pvl, Immune Evasion Cluster [sea, sep, chip, sak and scn] genes and biofilm operon icaABCD. On the other hand, multilocus sequence typing and agr typing methods were performed for the determination of clonal relationship and van operon was detected and sequenced.
Results: Vancomycin-resistant Staphylococcus aureus strain 1 (VRSA-1) was positive for vanA, ermA, ermC, aph(2”)-Ic, aph(3ʹ)-IIIa, sea, sep, icaD genes, belonging to agr type I; SCCmec type III; spa type t030; and ST239. However, the genetic characterization of Vancomycin-resistant Staphylococcus aureus strain 2 (VRSA-2) revealed the presence of various types of resistance genes vanA, ermA, ermC, aph(2”)-Ic, aph(3ʹ)-IIIa, sea, icaD, relating to agr type I; SCCmec type III; spa type t459; and ST239. The presence of transposon Tn1546 was determined by PCR sequencing.The Basic Local Alignment Search Tool analysis of van operon in the VRSA isolates showed 99.6% sequence homology to Tn1546 in vancomycin-resistant enterococci, indicating the vanA operon has an enterococcal origin.
Conclusion: In conclusion, the ST239 is one of the most common clones of MRSA isolates which involved the hospital-associated infections, therefore, the emergence of VRSA isolates with ST239 increased the spread of resistance to vancomycin in the hospital settings.
Keywords: VRSA, MLST, SCCmec, spa, agr type, van operon
Introduction
Staphylococcus aureus is one of the most destructive causes of bacterial infections in hospital settings.1 The rapid spread of this type of bacterial infections has been accompanied by a rise in antibiotic-resistant strains.2 The high-level vancomycin-resistant S. aureus (VRSA) was reported in 2002, Michigan, USA, for the first time and was subsequently detected in different hospitals around the world.3,4 However, VRSA strains are rarely reported from all over the world.5 So far, the VRSA strains have been reported, belonging to sequence types (STs) including ST1, ST5, ST8 ST85, ST231, ST239, ST250, ST371, ST923, and ST1283, and ST231.5–7 The VRSA strains belonging to ST5, ST8, ST239 and ST1283 were recently reported in Iran.6,7 The sequence type 239 is the most common clone of methicillin-resistant S. aureus (MRSA) which has been reported in the hospital settings around the world that would be resistant to the widespread antibacterial agents.8,9
Transposon Tn1546, a Tn3-related transposon,causing vancomycin resistance, was associated with a cluster of seven genes including vanS, vanR, vanH, vanA, vanX, vanY, and vanZ.10 Generally, the horizontal transfer of Tn1546 transposon from vancomycin-resistant Enterococci spp to S. aureus strains is responsible for resistance to vancomycin in S. aureus strains as well as the emergence of VRSA.5 Herein, we reported the genetic characterization of two vancomycin-resistant S. aureus strains which were collected from hospitalized patients, Kerman, Iran.
Materials and methods
Patients and isolates sources
In our previous studies, from April 2015 to March 2017, we detected two VRSA strains from 205 non-duplicate of S. aureus isolates. The two VRSA isolates were collected, VRSA-1 belonged to spa type t030; SCCmec III and VRSA-2 belonged to spa type t459; SCCmec III. The minimum inhibitory concentration (MIC) of isolates to vancomycin was ≥64 µg/mL and the isolates were negative for panton-valentine leukocidin (pvl) gene and were resistant to gentamicin, amikacin, erythromycin, clindamycin, tetracycline, ciprofloxacin, penicillin, cefoxitin, and trimethoprim/sulfamethoxazole11,12 The isolates were stored in Trypticase Soy Broth plus 15% Glycerol at −80°C for more investigation.
The VRSA strains were isolated from two patients with a long history of hospitalization. Briefly, the VRSA-1 was associated with the bronchoalveolar lavage sample from a 76-year-old female patient, hospitalizing in ICU at Afzalipour hospital (425 beds) in Kerman along with clinical manifestations including pneumonia, fever, with a history of diabetes mellitus and hemodialysis,who died without including infections treated. The VRSA-2 isolate was obtained from a surgical wound infection of a 21-year-old male patient, hospitalized in orthopedic unit in Bahonar hospital (375 beds) in Kerman. (Other data about patient was not available.)
Antibiotic susceptibility test
The MIC of the isolates to linezolid and daptomycin was determined according to Clinical & Laboratory Standards Institute guidelines and also, disk diffusion method was used for determination of susceptibility of isolates to rifampin.13
Detection of virulence, resistance genes, and molecular typing of the VRSA isolates
In the following previous studies,11,12 the isolates were confirmed by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF MS, Bruker, Daltonics, Medical Microbiology and Clinical Microbilogy Department of Umea University, Umea, Sweden). Resistance genes including ermA, ermB, ermC, msrA/B, aminoglycoside modifying genes [(aac(6ʹ)-Ie-aph(2ʹ’)-I, aph(2ʹ’)-Ib, aph(2ʹ’)-Ic, aph(2ʹ’)-Id, aph(3ʹ)-IIIa, ant(4ʹ)-Ia)], and agr type of the isolates were determined by PCR method in FlexCycler2 PCR-Thermocycler (Analytik Jena, Co, Jena, Germany). On the other hand, the virulence genes including sea, sep, chip, sak, scn, icaABCD were detected by PCR. Table 1 shows the primers that were used in PCR amplification of virulence, resistance genes, and agr typing. Multilocus sequence typing was performed by PCR sequencing of seven housekeeping genes (arc, aro, glp, gmk, pta, tpi, and yqi) according to https://pubmlst.org/saureus/.
 |
Table 1 The primer sequences were used in this study |
Detection and sequencing of van operon
The van operon including vanR, vanS, vanH, vanA, vanX, vanY, and vanZ were detected and sequenced by primer walking method, using 12 specific primer pairs (Table 2) that were designed by primer designing tool in NCBI (https://www.ncbi.nlm.nih.gov/tools/primer-blast) for different regions of Tn1546 by PCR technique. We used the Tn1546 sequences with accession numbers HM565172.1, M97297, KR349520, KR047792, and KX496042.1 in GenBank for primer designing (https://www.ncbi.nlm.nih.gov/nuccore). The PCR products for van genes were sequenced by Applied Biosystems 3730/3730Xl DNA Analyzers (Bioneer, Co, South Korea). The sequences were assembled by Lasergene package software (DNASTAR) and were analyzed by the basic local alignment search tool (BLAST) in NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Open reading frames (ORFs) of van genes were determined by Open Reading Frame Finder software (https://www.ncbi.nlm.nih.gov/orffinder).
 |
Table 2 Primers used for amplification and sequencing of van operon |
Results
Genetic characterization of isolates
The isolates were resistant to rifampin and sensitive to linezolid and daptomycin with MIC ≤0.5 µg/mL (antibiotic resistance profile to other antibiotic is given in “Material and methods“ section). The expected molecular size 1030 bp product referring to the vanA gene was detected and sequenced in both VRSA strains as shown in Figure 1. The sequence presented 100% identity with vanA gene of vancomycin-resistant enterococci (VRE), on Tn1546 as indicated in GenBank database. Furthermore, the VRSA strains were analyzed for the presence of resistance and virulence genes. VRSA-1 was positive for ermA, ermC, aph (2”)-Ic, aph (3ʹ)-IIIa, sea, sep, icaD genes. The strain belonged to SCCmec III, agrI, spa type t030 and ST239. VRSA-2 was positive for ermA, ermC, aph (2”)-Ic, aph (3ʹ)-IIIa genes, as well as the virulence genes including sea, icaD were detected. The VRSA-2 belonged to SCCmec III, agrI, spa type t459, and ST239.
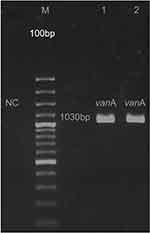 |
Figure 1 Agarose gel electrophoresis of vanA gene detected in two VRSA strains. |
Molecular characterization of van operon in VRSA strains
The amplification and sequencing of van operon showed that the operon contains 9 ORFs, harboring a transposase (tnp) and resolvase (rev) genes, corresponding to vanR, vanS, vanH, vanA, vanX, vanY, and vanZ operon, respectively. The analysis of the sequence results indicated that it is closely related to Tn1546 in VRE. Similar to Tn1546 sequence in VRE in the current work, the insertion sequences including IS1251 and IS1216E were also distinguished. Phylogenetic and distance analyses in BLAST revealed that Tn1546 in our VRSA strains had 99.6% similarity to Tn1546 in VRE. The sequence of Tn1546 was submitted in GenBank under accession number MG592387 (https://www.ncbi.nlm.nih.gov/nuccore/mg592387).
Discussion
Recently, due to the rapid prevalence of MRSA, reporting in health care settings and also the exposure of selective pressure of antibiotics, the emergence of VRSA strains, showing high level of resistance to antibiotics is unavoidable.
The emergence of VRSA isolates has been a major concern for the physicians and hospital management systems; however, VRSA isolates are rarely reported around the world and the total reported VRSA strains in the world have been reported to be less than 20 strains since 2002.5 Long-time hospitalization, prolonged treatment with vancomycin, colonization with enterococcal as well as MRSA isolates, diabetes, chronic skin ulcers, and hemodialysis are the risk factors for colonization and infection by VRSA.17,18 This study showed VRSA isolates from two patients who suffered from diabetes, chronic skin ulcers, underlying long-time hospitalization, and prolonged vancomycin therapy. The Centers for Disease Control and Prevention reported 9 out of 13 VRSA strains in the USA that were isolated from diabetic patients.17,18
The van operon is commonly located on transposon Tn1546 and causes high-level resistance to vancomycin, initially reported in hospital strains VRE.8,9 In the present study, both the isolates carried vanR, vanS, vanH, vanA, vanX, vanY, and vanZ genes on a van operon. Furthermore, we align the van genes sequence in BLAST, and results indicated high identity with Tn1546 in VRE. Therefore, it has been suggested that the van operon has probably been transferred from VRE to S. aureus strains in simultaneous infections or colonization of VRE along with S. aureus, although we did not distinguish any evidence of mixed infection or colonization with VRE in this study. In the present work, the VRSA as well as VRE strains were not isolated from roommates, other patients, and also health care workers. Furthermore, the presence of the IS elements including IS1251 and IS1216E in Tn1546 confirmed the transfer of resistance to vancomycin from VRE to S. aureus. Similar to present study, some VRSA strains were reported in the USA, harboring IS1251 and IS1216E on Tn1546.5,19 A study which was carried out in Kerman hospitals showed that vancomycin resistant-Enterococcus faecalis isolates harbored vanA gene.20
CC5 and CC8 are the most common MRSA clones around the world. ST239 from CC8, ST5 from CC5, and ST22 from CC22 are the most prevalent VRSA sequence types which were reported in Iran, USA, and Brazil.3–7 The sequence type (ST) 239 from CC8 is the most common clone of MRSA in hospital-acquired infections in Asia.7 According to literature, the ST239 is the epidemic clone with resistance to a wide range of antibacterial agents in hospital settings around the world.7 In recent years, the VRSA isolates belonging to ST5, ST8, ST239, and ST1283 were reported in our country.6,7 According to eBURST results, ST8 and also ST1283 are the single locus variant of ST239, and these findings indicated that, in our country, the VRSA isolates are closely related to the same clone (Figure 2). Also in the present study, the molecular typing results indicated that the genetic background of the VRSA isolates (agr type I; SCCmec type III; ST239 and pvl-negative isolates) was closely related to health-care-associated MRSA strains.
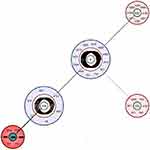 |
Figure 2 A population snapshot of the entire S. aureus with 5 or more nearest match of seven allelic profiles with ST239 in http://bigsdb.pasteur.fr MLST database showing clonal complexes (CCs) viewed using eBURST. |
ST239 is the common clone of the MRSA isolates that involved the hospital-associated infections especially in ICU-associated infections, and so the emergence of VRSA isolate, belonging to ST239 in health care settings, would be prevalent more than the other clones of MRSA (another clone of MRSA). Therefore, clinical laboratories have an important role in the diagnosis, isolation, and infection control with VRSA strains. So, we suggest that the surveillance of long-time hospitalized patients and the use of vancomycin agar screening for detecting VRSA and VISA strains in different hospital wards especially in ICU which probability of MRSA carriage and prolonged vancomycin therapy in them is high.
Ethical statement
The S. aureus strains were originally taken as part of routine hospital procedure, and then specifically recovered for this work. This study was approved by ethical numbers: IR.KMU.REC.1395.859 in ethical committee of Kerman University of Medical Sciences.
Acknowledgments
This research was supported by the research council and the student research committee of Kerman University of Medical Sciences, Kerman, Iran, and grant numbers: 94/677 and 95000434.
Author contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Chen CJ, Huang YC. New epidemiology of Staphylococcus aureus infection in Asia. Clin Microbiol Infect. 2014;20(7):605–623. doi:10.1111/1469-0691.12705
2. McGuinness WA, Malachowa N, DeLeo FR. Focus: infectious diseases: vancomycin resistance in Staphylococcus aureus. Yale J Biol Med. 2017;90(2):269–281.
3. Limbago BM, Kallen AJ, Zhu W, Eggers P, McDougal LK, Albrecht VS. Report of the 13th vancomycin-resistant Staphylococcus aureus isolate from the United States. J Clin Microbiol. 2014;52(3):998–1002. doi:10.1128/JCM.02187-13
4. Whitener CJ, Park SY, Browne FA, et al. Vancomycin-resistant Staphylococcus aureus in the absence of vancomycin exposure. Clin Infect Dis. 2004;38(8):1049–1055. doi:10.1086/382357
5. Rossi F, Diaz L, Wollam A, et al. Transferable vancomycin resistance in a community-associated MRSA lineage. N Engl J Med. 2014;370:1524–1531. doi:10.1056/NEJMoa1303359
6. Azimian A, Havaei SA, Fazeli H, et al. Genetic characterization of a vancomycin-resistant Staphylococcus aureus isolate from the respiratory tract of a patient in a University Hospital in Northeastern Iran. J Clin Microbiol. 2012;50(11):3581–3585. doi:10.1128/JCM.01727-12
7. Shekarabi M, Hajikhani B, Chirani AS, Fazeli M, Goudarzi M. Molecular characterization of vancomycin-resistant Staphylococcus aureus strains isolated from clinical samples: a three year study in Tehran, Iran. PLoS One. 2017;12(8):e0183607. doi:10.1371/journal.pone.0183607
8. Cha HY, Moon DC, Choi CH, et al. Prevalence of the ST239 clone of methicillin-resistant Staphylococcus aureus and differences in antimicrobial susceptibilities of ST239 and ST5 clones identified in a Korean hospital. J Clin Microbiol. 2005;43(8):3610–3614. doi:10.1128/JCM.43.8.3610-3614.2005
9. Liu Y, Wang H, Du N, et al. Molecular evidence for spread of two major methicillin-resistant Staphylococcus aureus clones with a unique geographic distribution in Chinese hospitals. Antimicrob Agents Chemother. 2009;53(2):512–518. doi:10.1128/AAC.00804-08
10. Simjee S, White DG, McDermott PF, et al. Characterization of Tn1546 in vancomycin-resistant Enterococcus faecium isolated from canine urinary tract infections: evidence of gene exchange between human and animal enterococci. J Clin Microbiol. 2002;40(12):4659–4665. doi:10.1128/jcm.40.12.4659-4665.2002
11. Fasihi Y, Saffari F, Mansouri S, Kalantar-Neyestanaki D. The emergence of vancomycin-resistant Staphylococcus aureus in an intensive care unit in Kerman, Iran. Wien Med Wochenschr. 2018;168(3–4):85–88. doi:10.1007/s10354-017-0562-6
12. Fasihi Y, Kiaei S, Kalantar-Neyestanaki D. Characterization of SCCmec and spa types of methicillin-resistant Staphylococcus aureus isolates from health-care and community-acquired infections in Kerman, Iran. J Epidemiol Glob Health. 2017;7(4):263–267. doi:10.1016/j.jegh.2017.08.004
13. CLSI. Performance Standards for Antimicrobial Susceptibility Testing. CLSI Supplement M100.
14. Emaneini M, Bigverdi R, Kalantar D, et al. Distribution of genes encoding tetracycline resistance and aminoglycoside modifying enzymes in Staphylococcus aureus strains isolated from a burn center. Ann Burns Fire Disasters. 2013;26(2):76–80.
15. Ahmadrajabi R, Layegh-Khavidaki SF, Kalantar-Neyestanaki D, Fasihi Y. Molecular analysis of Immune Evasion Cluster (IEC) genes and intercellular adhesion gene cluster (ica) among methicillin-resistant and methicillin-sensitive isolates of Staphylococcus aureus. J Prev Med Hyg. 2017;58(4):E308–E314. doi:10.15167/2421-4248/jpmh2017.58.4.711
16. Gilot P, Lina G, Cochard T, Poutrel B. Analysis of the genetic variability of genes encoding the RNA III-activating components Agr and TRAP in a population of Staphylococcus aureus strains isolated from cows with mastitis. J Clin Microbiol. 2002;40(11):4060–4067. doi:10.1128/jcm.40.11.4060-4067.2002
17. Centers for Disease Control and Prevention. CDC Reminds Clinical Laboratories and Healthcare Infection Preventionists of Their Role in the Search and Containment of Vancomycin-Resistant Staphylococcus Aureus (VRSA). Atlanta: CDC; 2012.
18. Cosgrove SE, Carroll KC, Perl TM. Staphylococcus aureus with reduced susceptibility to vancomycin. Clin Infect Dis. 2004;39:539–545. doi:10.1086/422458
19. Périchon B, Courvalin P. VanA-type vancomycin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2009;53(11):4580–4587. doi:10.1128/AAC.00346-09
20. Saffari F, Dalfardi MS, Mansouri S, Ahmadrajabi R. Survey for correlation between biofilm formation and virulence determinants in a collection of pathogenic and fecal Enterococcus faecalis isolates. Infect Chemother. 2017;49(3):176–183. doi:10.3947/ic.2017.49.3.176
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
