Back to Journals » Infection and Drug Resistance » Volume 15
Genetic Characterization of Enterobacter hormaechei Co-Harboring blaNDM-1 and mcr-9 Causing Upper Respiratory Tract Infection
Authors Liu H, Wang D, Tang M, Jia P, Huo Y, Wei E, Xu H, Chi X, Wang H
Received 19 March 2022
Accepted for publication 4 August 2022
Published 31 August 2022 Volume 2022:15 Pages 5035—5042
DOI https://doi.org/10.2147/IDR.S367073
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Huiqiong Liu,1 Dao Wang,1 Miaomiao Tang,1 Peisheng Jia,1 Yufeng Huo,1 Erhu Wei,1 Hao Xu,2 Xiaohui Chi,2 Huaili Wang1
1Department of Pediatric Intensive Care Unit, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, 450052, People’s Republic of China; 2Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, the First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, People’s Republic of China
Correspondence: Huaili Wang, Department of Pediatric Intensive Care Unit, The First Affiliated Hospital of Zhengzhou University, No. 1 Longhu East Zhonghuan Road, Zhengzhou, 450052, People’s Republic of China, Tel +86-371-66271057, Email [email protected]
Purpose: With the spread of multiple drug-resistant bacteria, blaNDM-1 and mcr-9 have been detected in various bacteria worldwide. However, the simultaneous detection of blaNDM-1 and mcr-9 in Enterobacter hormaechei has been rarely reported. This study identified an E. hormaechei strain carrying both blaNDM-1 and mcr-9. We investigated the genetic characteristics of these two resistance genes in detail, elucidating various potential mechanisms by which they may be transmitted.
Methods: Bacterial genomic features and possible origins were assessed by whole-genome sequencing (WGS) with Illumina and PacBio platforms and phylogenetic analysis. Subsequent investigations were performed, including antimicrobial susceptibility testing and multilocus sequence typing (MLST).
Results: We isolated an E. hormaechei strain DY1901 carrying both blaNDM-1 and mcr-9 from the sputum sample. Susceptibility testing showed that the isolate was multidrug-resistant. Multiple antibiotic resistance genes and virulence genes are widely distributed in DY1901. S1-PFGE, Southern blotting, and plasmid replicon typing showed that DY1901 carried four plasmids. The plasmid carrying mcr-9 was 259Kb in size and belonged to IncHI2, while the plasmid carrying blaNDM-1 was 45Kb in length and belonged to IncX3.
Conclusion: The E. hormaechei strain isolated in this study has a broad antibiotic resistance spectrum, posing a challenge to clinical treatment. Plasmids carrying mcr-9 are fusion plasmids, and those taking NDM are widely disseminated in China, suggesting that we should conduct routine genomic surveillance on such plasmids to curb the spread of drug-resistant bacteria in the region.
Keywords: E. hormaechei, New Delhi metallo-β-lactamase, mcr-9, whole-genome sequencing, phylogenetic analysis
Introduction
Enterobacter cloacae complex (ECC) comprises the following species: Enterobacter cloacae, Enterobacter hormaechei, Enterobacter asburiae, Enterobacter kobei Enterobacter ludwigii, Enterobacter nimipressuralis, Enterobacter mori, etc. ECC is a critical member of Enterobacteriaceae widely encountered in the environment.1,2 As an opportunistic pathogen, ECC is ubiquitous not only in nature but also in clinical settings and has been associated with various infections, such as bacteremia, respiratory tract infections, wound infections, and urinary tract infections.3,4
The prevalence of carbapenemase-resistant Enterobacteriaceae (CRE) has risen since the 2000s.5 New Delhi Metallo-β-lactamase (NDM) is a type of Metallo-β-lactamase (MBL) able to hydrolyze most β-lactams (including carbapenems).6,7 Colistin is an antibiotic often referred to as a “last resort” to treat CRE infections.8 The identification of the first mobile colistin resistance (MCR) gene, mcr-1, in 2015 triggered a rash of mcr screening reports.9 Subsequently, ten MCR-family genes and their variants have been described.10 Among the mcr-like genes, mcr-1 and mcr-9 are the most widespread. The mcr-9 gene has been found in 40 countries on six continents.11
In recent years, mcr family have continued to emerge in CRE and spread widely among pathogens of animal and human origin,12 further increasing the public health burden of antimicrobial resistance.13 This study isolated a strain of E. hormaechei carrying both blaNDM-1 and mcr-9 from the sputum of a patient with an upper respiratory tract infection. Importantly, these antibiotic resistance genes were located on different plasmids, signaling the potential spread of pan-resistant bacteria. However, the genetic characterization of plasmids and bacteria encoding NDM-1 and mcr-9 remains unclear. In this study, we identified an E. hormaechei strain carrying both blaNDM-1 and mcr-9. We investigated the genetic characteristics of these two resistance genes in detail, elucidating various potential mechanisms by which they may be transmitted.
Materials and Methods
Sample Collection
An E. hormaechei strain DY1901 carrying both blaNDM-1 and mcr-9 was isolated from the sputum of an inpatient in a tertiary hospital in China. A 72-year-old patient was admitted to hospital with left heart failure. The collected sputum was smeared with Mueller-Hinton Agar (OXOID, UK) medium and cultured by suctioning the sputum with a sterile suction device. Bacterial species were identified using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF/MS) (Bruker, Bremen, Germany). In addition, High throughput ANI analysis is used to compare the whole genome sequencing results to reveal distinct species of ECC (Figure S1). As previously described, the isolates were further subjected to PCR to detect the mcr and carbapenemase genes (Table S1).14
Antibiotic Susceptibility Testing
Minimum inhibitory concentrations (MICs) were determined by VITEK 2 system.15 The resistance results of tigecycline and colistin were interpreted following the European Committee on Antimicrobial Susceptibility Testing (EUCAST v12.0) guidelines. Susceptibilities against other drugs were determined by the criteria of the Clinical and Laboratory Standards Institute (CLSI v31). All methods were carried out under relevant guidelines and regulations.
Conjugation Assay and Plasmid Characterization
Conjugation experiments used E. coli J53 and E. coli EC600 as recipient strains. The resulting transformants were selected on BHI agar plates amended with meropenem (two mg/L). PCR and sequencing also screened the presence of blaNDM-1 and mcr-9. Plasmid sizes were determined using the S1-nuclease PFGE (S1-PFGE) method.16 According to the manufacturer’s instructions, southern blot hybridizations of plasmid DNA were performed on isolates using DIG-labelled probes (Roche Diagnostics, Germany).
Whole-Genome Sequencing
Whole-genome sequencing (WGS) was performed by Novo Gene Co., Ltd, Beijing, China. Genomic DNA was extracted using a DNA kit (Omega Bio-tek, Norcross, USA). The DNA was subsequently sequenced using Illumina-HiSeq 4000-PE150 (Illumina, San Diego, CA, USA) and PacBio RS II platform (Pacific Biosciences, California, USA). Alignment of antimicrobial resistance genes was performed through the ResFinder platform (https://cge.cbs.dtu.dk/services/ResFinder/). Using the website, multilocus sequence typing (MLST) was performed on all isolates (https://cge.cbs.dtu.dk/services/MLST/). The MLST of DY1901 was determined by aligning the housekeeping genes dnaA, fusA, gyrB, leuS, pyrG, rplB and rpoB. Virulence genes were identified by BLASTN against the VFDB database (http://www.mgc.ac.cn/VFs/main.htm).
Phylogenetic Reconstruction and Analysis
Complete E. hormaechei genomes were downloaded from the National Center for Biotechnology Information (NCBI) for phylogenetic analysis. Snippy (rapid haploid variant calling and core genome alignment) was used to compare genomic differences between strains. The alignment file was filtered from variants with elevated densities of base substitutions as putative recombination events by Gubbins version 2.4.1.17 The filtered core-genome alignment file was used to construct a maximum likelihood tree using FastTree with the GTR+CAT model.
Results
Antibiotic Resistance Signature of E. hormaechei
MALDI-TOF/MS and ANI analysis showed that DY1901 belonged to E. hormaechei (Figure S1), and then MIC was determined for E. hormaechei. The results of the MIC are shown in Table 1. DY1901 exhibited resistance to piperacillin/tazobactam, ceftazidime, ceftriaxone, cefepime, ertapenem, imipenem, levofloxacin, tetracycline, trimethoprim/sulfamethoxazole, but still susceptible to colistin, amikacin, and tigecycline. DY1901 has a broad drug resistance spectrum and is a multidrug-resistant bacterium.
 |
Table 1 Antibiotic Susceptibility Profiles of Enterobacter hormaechei Isolate DY1901 |
Antimicrobial Resistance Genes
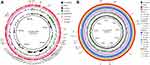
We provide data on antimicrobial resistance genes in Figure 1. DY1901 carries carbapenem resistance gene blaNDM-1 and colistin resistance gene mcr-9. In addition, DY1901 also carries β-lactams, sulfonamides, aminoglycosides, tetracyclines, and other antibiotic resistance genes (Figure 1). We also compared the resistance gene profiles from 30 E. hormaechei strains downloaded from NCBI. The results showed that the antibiotic resistance gene carried by 30 E. hormaechei strains was dominated by β-lactam resistance genes (93.6%), followed by quinolone resistance genes (Figure 1).
Virulence Genes
The results of the alignment of virulence genes are shown in Figure 1. The virulence genes carried by DY1901 are related to acriflavine resistance B (AcrB), type VI secretion system protein (T6SS), and transcriptional regulator RcsB. The 30 E. hormaechei strains downloaded from NCBI mainly encoded AcrB.
Molecular Characteristics of E. hormaechei
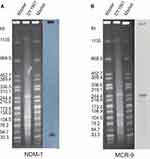
Detailed genomic analysis of E. hormaechei indicated that it belonged to ST418. The results of S1-PFGE, Southern blotting, and plasmid replicon typing showed that DY1901 carried four plasmids (Figure 2), the plasmid carrying mcr-9 was 259Kb in size and belonged to IncHI2, while the plasmid harboring blaNDM-1 was 45Kb in length and belonged to IncX3 (Figure 3). After annotating the genes on the plasmid, we found that the mcr-9 gene is located on the IS1R transposon element of pDY1901-mcr. Phylogenetic analysis of all 31 strains showed that DY1901 is closely related to GCF_003031755 and GCF_003339765 from the United States.
Discussion
The bacterial natural product colistin is considered the last line of defense against many Gram-negative pathogens,18 but with the spread of multiple drug-resistant bacteria, blaNDM-1 and mcr-9 have been detected in various bacteria worldwide.19–21 However, few reports have performed a detailed genomic analysis of E. hormaechei with blaNDM-1 and mcr-9.22–24 In this study, an E. hormaechei strain carrying both blaNDM-1 and mcr-9 was isolated from a patient. The isolate was characterized by genome and phylogenetic analysis using whole-genome sequencing technology.
By comparison, we found that pDY1901-mcr belonged to the IncHI2/IncHI2A plasmid, and no matching plasmid was found by NCBI, indicating that this plasmid may be a new hybrid plasmid. After annotating the genes on the plasmids, we performed a genetic environment analysis of mcr-9. It was found that the mcr-9 gene is located on the IS1R transposon element in pDY1901-mcr. IS1R transposon is relatively common in Klebsiella pneumoniae and Escherichia coli and is an essential reason for the spread of drug-resistant bacteria.25,26 In addition, we found that pDY1901-NDM belongs to the IncX3 plasmid and matched with the NDM-producing K. pneumoniae plasmid pCP028786.2, E. coli pCP050161, Salmonella pMH105050.1, ECC pMN061454.1. It is worth noting that these plasmids were all isolated from China, indicating that such plasmids have spread widely in China. After annotating the genes on the plasmids, we performed a genetic environment analysis of the NDM. The NDM gene was located on the IS5 transposon element in pDY1901-NDM, and the IS5 transposon significantly contributed significantly to the plasmid’s spread27 (Figure 3).
Bacterial resistance studies showed that DY1901 was resistant to piperacillin/tazobactam, ceftazidime, ceftriaxone, cefepime, ertapenem, imipenem, levofloxacin, tetracycline, trimethoprim/sulfamethoxazole. However, it was sensitive to colistin, amikacin, and tigecycline.28 The full expression of wild-type mcr-9 requires additional factors or inducing/derepression conditions, and promoter variation also affects mcr-9 expression, which may explain why the E. hormaechei queried here is sensitive to colistin under the tested conditions.29 A previous study showed that 48 E. hormaechei isolates were collected from a hospital in China from 2000 to 2018. The survey carried out MIC determination on 10 E. hormaechei isolates, and the results showed that all 10 E. hormaechei isolates were MDR. Similar to this study, E. hormaechei was only sensitive to colistin, tigecycline and some carbapenems. In response to the MIC results of DY1901, the patient was treated with tigecycline and E. hormaechei was not isolated from the patient’s sputum.
As the pathogen continued to be isolated, its resistance spectrum has changed, most notably due to genes encoding β-lactamase, leading to increased resistance to β-lactam antimicrobials.30 Antibiotic resistance genes can make antibiotics ineffective by changing the antibiotic molecular structure and antibiotic target protein.31 DY1901 in this study was resistant to ertapenem and imipenem. However, whole-genome sequencing showed that DY1901 carried blaNDM-1, indicating that the genotype was consistent with the bacterial resistance phenotype. In addition, the strain also carried antibiotic resistance genes such as β-lactams, sulfonamides, aminoglycosides, and tetracyclines and had multidrug-resistant phenotypes. Compared with the 30 isolates of E. hormaechei downloaded from NCBI, DY1901 lacks quinolone resistance genes, but DY1901 shows resistance to levofloxacin, which may be due to other resistance mechanisms of the pathogen.
Existing reports on E. hormaechei indicate that many E. hormaechei has a characteristic of strong drug resistance but weak virulence.32 The virulence genes carried by DY1901 in this study included AcrB, T6SS, and RcsB. T6SS belongs to the bacterial secretion system and is associated with bacterial colonization, and can help bacteria to transfer effector proteins into host cells or the extracellular matrix for a variety of purposes, including bacterial persistence, adhesion, virulence, invasion, biofilms Formation, and inter-bacterial competition.33,34 AcrB belongs to the multidrug efflux systems of bacteria, and bacteria are intrinsically resistant to cytotoxic substances through their network of outer membranes and multidrug efflux systems, acting synergistically, thereby reducing the impact of antibiotics on it.35,36
Phylogenetic analysis of DY1901 and the complete genomes of E. hormaechei in NCBI showed that E. hormaechei mainly had three clones, and DY1901 was located in the second clone (Figure 1). From the evolutionary tree, it can be found that DY1901, GCF_003031755, and GCF_003339765 have the closest relationship, so we speculate that DY1901 may have evolved from the strain in the United States. This result reflects the global nature of bacterial resistance from another perspective, which requires extensive attention from worldwide.
Conclusions
We report the detection of an E. hormaechei strain carrying both blaNDM-1 and mcr-9. This study demonstrated the genetic characterization of plasmids carrying blaNDM-1 and mcr-9 by whole-genome sequencing. The E. hormaechei strain isolated in this study has a broad drug resistance spectrum, posing a challenge to clinical treatment. Plasmids carrying mcr-9 are fusion plasmids, and those carrying NDM are widely disseminated in China. This suggests that we should conduct routine genomic surveillance on such plasmids to effectively curb the spread of drug-resistant bacteria in the region.
Data Sharing Statement
The whole-genome sequences of the E. hormaechei were submitted to GenBank under the following BioProject numbers: PRJNA808678.
Ethical Statement
Written informed consents were obtained from patients. This study was conducted following the Declaration of Helsinki and obtained approval from the Medical Ethics Committee at The First Affiliated Hospital of Zhengzhou University. The ethics permit number is KY-2022-0379.
Funding
This project was supported by the Henan Provincial Key Scientific and Technological Research Program (SBGJ202002114).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Sanders WE, Sanders CC. Enterobacter spp.: pathogens poised to flourish at the turn of the century. Clin Microbiol Rev. 1997;10(2):220–241. doi:10.1128/CMR.10.2.220
2. Mezzatesta ML, Gona F, Stefani S. Enterobacter cloacae complex: clinical impact and emerging antibiotic resistance. Future Microbiol. 2012;7(7):887–902. doi:10.2217/fmb.12.61
3. Chen J, Tian S, Nian H, et al. Carbapenem-resistant Enterobacter cloacae complex in a tertiary Hospital in Northeast China, 2010–2019. BMC Infect Dis. 2021;21(1):611. doi:10.1186/s12879-021-06250-0
4. Huang J, Xu Q, Liu F, Xiong H, Yang J. Enterobacter cloacae infection of the shoulder in a 52-year-old woman without apparent predisposing risk factor: a case report and literature review. BMC Infect Dis. 2021;21(1):13. doi:10.1186/s12879-020-05699-9
5. Munoz-Price LS, Poirel L, Bonomo RA, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13(9):785–796.
6. Nordmann P, Poirel L, Walsh TR, Livermore DM. The emerging NDM carbapenemases. Trends Microbiol. 2011;19(12):588–595. doi:10.1016/j.tim.2011.09.005
7. Han R, Shi Q, Wu S, et al. Dissemination of carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) among carbapenem-resistant enterobacteriaceae isolated from adult and children patients in China. Front Cell Infect Microbiol. 2020;10:314. doi:10.3389/fcimb.2020.00314
8. Li J, Nation RL, Turnidge JD, et al. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect Dis. 2006;6(9):589–601. doi:10.1016/S1473-3099(06)70580-1
9. Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi:10.1016/S1473-3099(15)00424-7
10. Bitar I, Papagiannitsis CC, Kraftova L, Chudejova K, Mattioni Marchetti V, Hrabak J. Detection of five mcr-9-carrying enterobacterales isolates in four Czech hospitals. mSphere. 2020;5(6). doi:10.1128/mSphere.01008-20
11. Li Y, Dai X, Zeng J, Gao Y, Zhang Z, Zhang L. Characterization of the global distribution and diversified plasmid reservoirs of the colistin resistance gene mcr-9. Sci Rep. 2020;10(1):8113. doi:10.1038/s41598-020-65106-w
12. Lu X, Du Y, Peng K, et al. Coexistence of tet(X4), mcr-1, and blaNDM-5 in ST6775 Escherichia coli Isolates of Animal Origin in China. Microbiol Spectr. 2022;10(2):e0019622. doi:10.1128/spectrum.00196-22
13. Wang X, Wang Y, Zhou Y, et al. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg Microbes Infect. 2018;7(1):122. doi:10.1038/s41426-018-0124-z
14. Zheng B, Zhang J, Ji J, et al. Emergence of Raoultella ornithinolytica coproducing IMP-4 and KPC-2 carbapenemases in China. Antimicrob Agents Chemother. 2015;59(11):7086–7089. doi:10.1128/AAC.01363-15
15. Lellouche J, Schwartz D, Elmalech N, et al. Combining VITEK((R)) 2 with colistin agar dilution screening assist timely reporting of colistin susceptibility. Clin Microbiol Infect. 2019;25(6):711–716. doi:10.1016/j.cmi.2018.09.014
16. Xu H, Wang X, Yu X, et al. First detection and genomics analysis of KPC-2-producing Citrobacter isolates from river sediments. Environ Pollut. 2018;235:931–937. doi:10.1016/j.envpol.2017.12.084
17. Croucher NJ, Page AJ, Connor TR, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43(3):e15. doi:10.1093/nar/gku1196
18. Wang Z, Koirala B, Hernandez Y, et al. A naturally inspired antibiotic to target multidrug-resistant pathogens. Nature. 2022;601(7894):606–611. doi:10.1038/s41586-021-04264-x
19. Kim JS, Jin YH, Park SH, et al. Horizontal transfer of blaNDM-1-carrying IncX3 plasmid between carbapenem-resistant Enterobacteriaceae in a single patient. J Infect. 2020;81(5):816–846. doi:10.1016/j.jinf.2020.07.013
20. Zheng B, Dong H, Xu H, et al. Coexistence of MCR-1 and NDM-1 in clinical Escherichia coli isolates. Clin Infect Dis. 2016;63(10):1393–U1145. doi:10.1093/cid/ciw553
21. Osei Sekyere J, Maningi NE, Modipane L, Mbelle NM. Emergence of mcr-9.1 in extended-spectrum-beta-lactamase-producing clinical Enterobacteriaceae in Pretoria, South Africa: global evolutionary phylogenomics, resistome, and mobilome. mSystems. 2020;5(3). doi:10.1128/mSystems.00148-20
22. Lin M, Yang Y, Yang Y, et al. Co-occurrence of mcr-9 and bla NDM-1 in Enterobacter cloacae isolated from a patient with bloodstream infection. Infect Drug Resist. 2020;13:1397–1402. doi:10.2147/IDR.S248342
23. Ai W, Zhou Y, Wang B, et al. First report of coexistence of bla SFO-1 and bla NDM-1 beta-lactamase genes as well as colistin resistance gene mcr-9 in a transferrable plasmid of a clinical isolate of Enterobacter hormaechei. Front Microbiol. 2021;12:676113. doi:10.3389/fmicb.2021.676113
24. Sun L, Zhao X, Wang L, Guo X, Shi X, Hu L. Coexistence of mcr-9 and blaNDM-1 in a multidrug-resistant Enterobacter hormaechei strain recovered from a bloodstream infection in China. J Glob Antimicrob Resist. 2021;24:440–442. doi:10.1016/j.jgar.2021.02.011
25. Beyrouthy R, Robin F, Delmas J, et al. IS1R-mediated plasticity of IncL/M plasmids leads to the insertion of bla OXA-48 into the Escherichia coli chromosome. Antimicrob Agents Chemother. 2014;58(7):3785–3790. doi:10.1128/AAC.02669-14
26. Lev AI, Astashkin EI, Shaikhutdinova RZ, et al. Identification of IS1R and IS10R elements inserted into ompk36 porin gene of two multidrug-resistant Klebsiella pneumoniae hospital strains. FEMS Microbiol Lett. 2017;364(10). doi:10.1093/femsle/fnx072
27. Uz Zaman T, Albladi M, Siddique MI, Aljohani SM, Balkhy HH. Insertion element mediated mgrB disruption and presence of ISKpn28 in colistin-resistant Klebsiella pneumoniae isolates from Saudi Arabia. Infect Drug Resist. 2018;11:1183–1187. doi:10.2147/IDR.S161146
28. Iwu CD, du Plessis EM, Korsten L, Nontongana N, Okoh AI. Antibiogram signatures of some enterobacteria recovered from irrigation water and agricultural soil in two district municipalities of South Africa. Microorganisms. 2020;8(8):1206. doi:10.3390/microorganisms8081206
29. Carroll LM, Gaballa A, Guldimann C, Sullivan G, Henderson LO, Wiedmann M. Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype typhimurium isolate. mBio. 2019;10(3). doi:10.1128/mBio.00853-19
30. Cabral AB, Maciel MAV, Barros JF, Antunes MM, Barbosa de Castro CMM, Lopes ACS. Clonal spread and accumulation of beta-lactam resistance determinants in Enterobacter aerogenes and Enterobacter cloacae complex isolates from infection and colonization in patients at a public hospital in Recife, Pernambuco, Brazil. J Med Microbiol. 2017;66(1):70–77. doi:10.1099/jmm.0.000398
31. Christaki E, Marcou M, Tofarides A. Antimicrobial resistance in bacteria: mechanisms, evolution, and persistence. J Mol Evol. 2020;88(1):26–40. doi:10.1007/s00239-019-09914-3
32. Davin-Regli A, Lavigne JP, Pages JM. Enterobacter spp.: update on taxonomy, clinical aspects, and emerging antimicrobial resistance. Clin Microbiol Rev. 2019;32(4). doi:10.1128/CMR.00002-19
33. Soria-Bustos J, Ares MA, Gomez-Aldapa CA, Gonzalez YM, Giron JA, De la Cruz MA. Two type VI secretion systems of enterobacter cloacae are required for bacterial competition, cell adherence, and intestinal colonization. Front Microbiol. 2020;11:560488. doi:10.3389/fmicb.2020.560488
34. Storey D, McNally A, Astrand M, et al. Klebsiella pneumoniae type VI secretion system-mediated microbial competition is PhoPQ controlled and reactive oxygen species dependent. PLoS Pathog. 2020;16(3):e1007969. doi:10.1371/journal.ppat.1007969
35. Kobylka J, Kuth MS, Muller RT, Geertsma ER, Pos KM. AcrB: a mean, keen, drug efflux machine. Ann N Y Acad Sci. 2020;1459(1):38–68. doi:10.1111/nyas.14239
36. Zhang P, Mao D, Gao H, et al. Colonization of gut microbiota by plasmid-carrying bacteria is facilitated by evolutionary adaptation to antibiotic treatment. ISME J. 2021;16:1284–1293.
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.