Back to Journals » Infection and Drug Resistance » Volume 12
Genetic basis for metronidazole and clarithromycin resistance in Helicobacter pylori strains isolated from patients with gastroduodenal disorders
Authors Hashemi SJ , Sheikh AF, Goodarzi H, Yadyad MJ, Seyedian SS, Aslani S , Assarehzadegan MA
Received 31 October 2018
Accepted for publication 22 January 2019
Published 4 March 2019 Volume 2019:12 Pages 535—543
DOI https://doi.org/10.2147/IDR.S192942
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Seyed Jalal Hashemi,1–3 Ahmad Farajzadeh Sheikh,1,4 Hamed Goodarzi,1,4,5 Mohammad Jaafar Yadyad,1 Seyed Saeid Seyedian,6 Sajad Aslani,7 Mohammad-Ali Assarzadegan8
1Infectious and Tropical Diseases Research Center, Health Research Institute, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran; 2Research Institute for Infectious Diseases of the Digestive System, School of Medicine, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran; 3Division of Gastroenterology and Hepatology, Ahvaz Jundishapur University of Medical Sciences, Ahwaz, Iran; 4Department of Microbiology, School of Medicine, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran; 5Molecular Biology Research Center, Baqiyatallah University of Medical Science, Tehran, Iran; 6Alimentary Tract Research Center, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran; 7Student Research Committee, Kerman University of Medical Sciences, Kerman, Iran; 8Immunology Department, School of Medicine, Iran University of Medical Sciences, Tehran, Iran
Background: The aim of this study was to evaluate the antimicrobial resistance and genetic basis for metronidazole (Mtz) and clarithromycin (Cla) resistance in strains of Helicobacter pylori, isolated from patients with gastroduodenal disorders.
Patients and methods: A total of 157 H. pylori isolates (from 22 gastric cancer, 38 peptic ulcer disease, and 97 non-ulcer dyspepsia patients) were analyzed for drug susceptibility to Mtz and Cla, by gradient diffusion test (E-test, MAST). The PCR and sequence analysis of the rdxA and frxA for Mtz-resistant strains and the 23S rRNA for Cla-resistant strains were used to determine the genetic basis of drug resistance in H. pylori strains. Increased expression of TolC homologous genes (hefA) that upregulates efflux pump activity was determined in multidrug-resistant (MDR) strain of H. pylori by real-time PCR technique.
Results: Among 157 H. pylori isolates, 32 (20.4%) strains were resistant to at least one of the antimicrobial agents. The highest resistance rate was attributed to Mtz (n=69, 43.94%). Among the resistant strains of H. pylori, 15 cases (9.55%) were detected as MDR. Mutations in the rdxA (85.5%) and A2143G point mutations (63.1%) in the 23S rRNA were the most common cause of resistance to Mtz and Cla in strains of H. pylori, respectively. In MDR strains, the rdxA mutation and A2143G-point mutation in the 23S rRNA were the most abundant mutations responsible for drug resistance. The relative expression of hefA in MDR strains (mean 3.706) was higher than the susceptible strains (mean 1.07).
Conclusion: Mutational inactivation and efflux pump overexpression are two mechanisms that increase the resistance to H. pylori antimicrobial agents and the rate of MDR strains. In Iran, the mutations of rdxA and frxA in Mtz-resistant strains and A2143G and A2142G of the 23S rRNA in Cla-resistant strains were significant. The screening for these mutations could help to prevent antibiotic resistance, and to determine the most effective anti-H. pylori drugs.
Keywords: H. pylori, drug resistance, efflux pump, genetic mutations
Introduction
Helicobacter pylori is an etiological factor for gastroduodenal disorders such as chronic gastritis, peptic ulcer disease (PUD), and in more severe cases, H. pylori infection may result in gastric cancer (GC).1 Treatment of H. pylori infection is usually a challenge because the strains of H. pylori are susceptible to most antimicrobial agents in vitro, but in vivo the successful treatment is hard.2 Treatment process is usually performed by a combined therapy regimen that recommended eradicating the H. pylori infection. The emergence of antibiotic resistance is considered as the major cause of treatment failure.3,4 A successful combination therapy of a proton pump inhibitor and simultaneous prescription of two or three kinds of antibiotics, including tetracycline (Tet), metronidazole (Mtz), clarithromycin (Cla), and amoxicillin (Amx) are recommended for treatment of H. pylori infection. The prevalence of antibiotic resistance of strains of H. pylori varies in different geographical regions, and is associated with the consumption of antibiotics in those areas.5 In recent years, the widespread use of antibiotics has increased the rate of resistance of H. pylori to standard therapies. Resistance to Mtz and Cla has increased worldwide, and also multidrug-resistant (MDR) strains of H. pylori and simultaneous resistant to Mtz, Cla, and Amx have been reported.6,7 Recent studies suggest that the underlying antibiotic resistance mechanisms are mainly due to the genetic plasticity of H. pylori that results in genetic mutations.7–9 These include point mutation, involving substitution of A to G at positions 2142 and 2143, in the domain V of the 23S rRNA, which confers resistance toward Cla.9 Similarly, mutations in the rdxA and frxA are reported to confer Mtz resistance in H. pylori.9 Mutational patterns in the 23S rRNA, rdxA, and frxA are caused by the inactivation of the gene function via frameshift mutation, insertions, and deletions, and showed wide geographical divergence.
In addition, possible mechanisms of intrinsic drug resistance involve decreased drug uptake or increased drug efflux. Efflux of compounds is a phenomenon commonly observed in bacteria.8,10,11 Five families of multidrug efflux transporters have been described. One of them, widespread in Gram-negative bacteria, is the resistance-nodulation-division (RND) family of efflux systems.12–14 There are three RND families: hefABC, hefDEF, and hefGHI. The hefA, hefD, and hefG are the TolC homolog, encoding the outer membrane protein belonging to efflux pump. About 69% of the Iranian population were infected by H. pylori, and failure in treatment could be caused by the development of partial gastritis to PUD or GC.15 The aim of the present study was to investigate the frequency of drug resistance in H. pylori strains, and to employ the molecular methods for analysis of genes related to Mtz and Cla resistance. This study may have the potential to improve the prognosis and empiric treatment in Ahvaz, Iran, in a clinical setting.
Recent studies suggest that the underlying antibiotic resistance mechanisms are mainly due to the genetic plasticity of H. pylori that result in genetic mutations. These include point mutation, involving substitution of A to G at positions 2142 and 2143, in the domain V of the 23S rRNA, which confers resistance toward Cla.8 Similarly, mutations in the rdxA and frxA are reported to confer Mtz resistance in H. pylori.9 Unlike mutational patterns in the 23S rRNA, the mutations in the rdxA and frxA are caused by the inactivation of the gene function via frameshift mutation, insertions, and deletions.
Patients and methods
Ethics statement
The study was approved by the Research Ethics Committee of the Ahvaz Jundishapur University of Medical Sciences (No: IR.AJUMS.REC.1395.220.), Ahvaz, Iran. Written informed consent was obtained from all the gastroduodenal patients.
Bacterial culture and DNA isolation
A total 157 H. pylori (22 GC, 38 PUD, and 97 non-ulcer dyspepsia [NUD]) strains originally isolated between 2015 and 2016 from 150 patients living in Ahvaz, Khuzestan province, Iran, were selected. The patients who had not received nonsteroidal anti-inflammatory drugs or any anti-Helicobacter therapy at least for 3 months prior to endoscopy were included. Upper gastro-endoscopy was performed on patients’ with clinical symptoms, including gastritis, abdominal pain, weight loss, epigastric pain, vomiting, dyspepsia and history of GC, melena, anemia, and history of gastric ulcer for biopsy collection. Brucella agar (EMD Millipore, Billerica, MA, USA), supplemented with 7% defibrinated sheep blood, 7% fetal calf serum (FCS), 10 mg/L vancomycin, 5 mg/L amphotericin B, 5 mg/L trimethoprim, and 20 U/mL polymyxin B, was used to culture the H. pylori strains under microaerophilic conditions (10% CO2, 5% O2, and 85% N2; EMD Millipore) for 5–7 days, and all the plates were incubated at 37°C with 100% humidity. Based on colony morphology (small and translucent colonies), bacterial shape (curved), and a Gram-negative stain, and also using biochemical tests, including urease, catalase, and oxidase, preliminary detection of H. pylori was performed. After sub-culturing a colony (twice or thrice) on the Brucella agar, supplemented with 7% sheep blood, all the isolates were stored in skim milk with 20% glycerol and 7% FCS at −70°C.16,17 Genomic DNA was extracted from antral biopsy and the colonies using the high pure DNA extraction kit (Hoffman-La Roche Ltd., Basel, Switzerland), according to the manufacturer’s instruction. Then, the extracted DNA was quantified, using a biophotometer through measurement of the associated OD at 260 nm (Eppendorf, Hamburg, Germany).
Detection of H. pylori by PCR assay
For preliminary detection of H. pylori strains, the PCR assay was performed using the primers based on the H. pylori 16S rDNA.18 PCR assays were performed using one set of oligonucleotides (Hpx1/Hpx2) (5′-CTGGAGARACTAAGYCCTCC-3′ and 5′-GAG-GAATACTCATTGCGAAGGCGA-3′), amplifying a 150-bp fragment.19 PCR assay was carried out in a thermocycler nexus gradient (Eppendorf). A 25 µL final reaction, including 10× PCR buffer, 1.5 mM MgCl2, 10 mM dNTPs, 0.5 µM of each primer, 1.5 U super Taq polymerase, and 5 µL of template DNA was prepared for PCR assay. The cycling parameters were 40 cycles of 95°C for 5 minutes, 56°C for 1 minute, and 72°C for 1 minute. The amplified products were detected by electrophoresis on a 2% agarose gel containing ethidium bromide (500 ng/mL). The bands were visualized using A10 system (Bio-Rad Laboratories Inc., Hercules, CA, USA).
Antibiotic susceptibility testing
Bacterial colonies obtained during 72-hour culture were suspended in the brain heart infusion broth with a density of three on the McFarland scale, 108 cells CFU/mL. In the next stages, the bacterial suspension was inoculated on Mueller-Hinton medium (Becton Dickinson, Franklin Lakes, NJ, USA), supplemented with 10% defibrinated sheep blood. Then E-test antibiotic impregnated with a gradient of concentration was imposed to determine the value of minimum inhibitory concentration (MIC) for the growth of bacteria. Plates were then incubated at 37°C for 72 hours under microaerophilic conditions. Criteria for interpretation of results were MIC (µg/mL) for resistant strains: Amx ≥2, Cla ≥1, Mtz ≥8, Tet ≥4, and ciprofloxacin (Cip) ≥1. H. pylori strain ATCC43504 was used as a reference strain for the quality control of antibiotic susceptibility testing.20–23
Molecular detection on H. pylori-resistant strains
The PCR-based sequencing was used for genetic analysis of antibiotic-resistant strains. Genes related to drug resistance, including the rdxA, frxA, and 23S rRNA were amplified by PCR using specific primers as previously described.9,24,25 The PCR products were separated using a 2% agarose gel stained by safe stain and visualized under UV light. Target band was extracted by the GF-1 Gel DNA Recovery Kit (Vivantis Technologies Sdn. Bhd., Selangor Darul Ehsan, Malaysia).
Sequence data analysis
The target PCR products belonging to the rdxA, frxA, and 23S rRNA in 157 strains of H. pylori isolated from GC, PUD, and NUD patients were collected and sent for sequence analysis to the Bioneer Corporation, Daejeon, South Korea. Comparative sequence analysis between resistant and sensitive strains was performed by MEGA version 6 software.26 H. pylori 26695 (GenBank accession number NC_000915.1; GI: 15644634) was used as a reference genome for alignment of sequence data.
The hefA expression in MDR and the reference strain by real-time (RT)-PCR assay
Total RNA extraction
Fifteen isolates of MDR strains of H. pylori were cultured on Mueller-Hinton agar, supplemented with 5% defibrinated sheep blood, 7% FCS, 1, 2, and 8 µg/mL of Cla, Amx, and Mtz, respectively. All plates were incubated at 37°C, under microaerophilic conditions. Bacterial suspension was prepared in normal saline. The cells were then isolated by centrifugation (9,000×g for 10 minutes), and the pellet was used for total RNA extraction. RNA in MDR strains was isolated using the RNA extraction kit (Hoffman-La Roche Ltd.) and then reverse transcribed into cDNA. The hefA vs gyrB (an endogenous gene) was used to study the relative expression of the hefA in 15 MDR clinical strains and strain ST7136 of H. pylori. cDNA of hefA and gyrB was amplified using RT-PCR (ABI StepOne™; Thermo Fisher Scientific, Waltham, MA, USA) in the presence of Real Master Mix (SYBR Green; TaKaRa, Shiga, Japan). The sequences of hefA (GenBank, accession number AF059041), including F: (5′-CTCGCTCGCATGATCGC-3 and R: (5′-CGTATTCGCTCAAA-TTCCCT-3′) were used to amplify a 162-bp fragment. The expression of the housekeeping gene (GenBank, accession number AB084049) was assessed in parallel with the primer pair gyrB: F: (5′-TTACTACGACTTATCCTGGGGCTAGCGCTG-3′) and R: (5′-CCCCATCAAT-TTCCACATTCTCCGC-3′), amplifying a 267-bp fragment. Then, the hefA expression in MDR and susceptible strain were determined.27
Results
Assessment of susceptibilities to antimicrobial agents in clinical strains
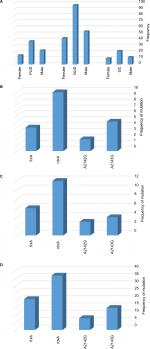
The MIC of Mtz and Cla was determined for 157 strains of H. pylori. Both biopsy samples and colonies were confirmed by PCR analysis in 22 GC, 38 PUD, and 97 NUD patients by gradient diffusion test. In general, among 157 isolates of H. pylori, 32 (20.4%) strains were resistant to only one of the antimicrobial agents (Table 1). Single resistance was observed only in Mtz or Cip. The highest resistance rate was attributed to Mtz (n=69, 43.94%). The frequency of double and MDR phenotypes is listed in Table 2. Among the resistant strains, MDR strains were detected in 3 (13.6%), 5 (13.2%), and 7 (7.2%) cases in GC, PUD, and NUD patients, respectively (Table 2).
Genetic variations of the rdxA and frxA in Mtz-resistant strains
The sequences of the rdxA and frxA were analyzed in 57 Mtz-resistant and 5 Mtz-sensitive strains of H. pylori (Figure 1). Sequences of the rdxA and frxA in sensitive strains were pairwise aligned with a reference strain by MEGA version 5 software, showing 95.6%–97.1% and 97.3%–98.7% identity in both genes, respectively. Alteration in amino acid sequences of the rdxA was detected in 50/57 (87.7%) Mtz-resistant strains. Out of 50 strains, 28 (49.1%) translational frameshifts were caused by the presence of premature stop codon through substitution of amino acid in the truncated rdxA (Table 3). The rdxA mutations, including missense and nucleotide insertion/deletion were detected in 8 (14%) and 14 (24.6%), respectively, in the Mtz-resistant strains. In addition, the premature stop codon (29.8%) was seen as the most common mutation in the frxA in Mtz-resistant strains (Table 3). In the MDR strain, the truncated rdxA and frxA were detected as the most common mutation responsible for Mtz resistance (Table 4).
Genetic variations of the 23S rRNA in Cla-resistant strains
The V domain of 23S rRNA was sequenced in the 38 Cla-resistant strains and compared to the reference genome of H. pylori 26695 exhibiting two-point mutations, including substitution of a single nucleotide A2143G (63.1%) and A2142G (34.2%) in 24 and 13 strains, respectively (Figure 1). Simultaneous double mutations of the A2143G and A2142G have not been found in these strains. The A2142C, A2144T, C2245T, G2141A, and G2224A were among the other mutations that have been found in Cla-resistant strains of H. pylori (Table 3).
Expression of efflux pump hefA gene in MDR strains
The expression of hefA and gyrB in MDR and susceptible strain was assessed by RT-PCR assay. Each relative expression value was the mean of three replicates. In 15 clinical isolates, the relative expression of hefA vs gyrB in MDR strains of H. pylori (mean 3.706) was higher than the seven susceptible strains (mean 1.07) (Table 5).
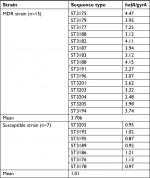  | Table 5 Relative expression of hefA gene vs gyrB in MDR clinical strains compared to susceptible strain Abbreviation: MDR, multidrug resistant. |
Discussion
In Iran, like other developing countries, H. pylori has infected the majority of the adult population. According to serology data, the incidence of H. pylori infection in Iranian adults is up to 80%.28 In the present study, resistance to Mtz, Cla, Tet, Amx, and Cip were determined in strains of H. pylori by gradient diffusion test (E-test, MAST). Single resistance was observed only in two antibiotics; Mtz (17.2%) and Cip (3.2%), while double resistance and MDR were seen in 11.5% and 20.4%, respectively. Among 157 H. pylori isolates, 82 (52.2%) cases were resistant to at least one antibiotic, and the highest resistance rate was attributed to Mtz (n=69, 43.9%). The prevalence of resistance to Mtz in strains of H. pylori in Iran is high; the prescription of Mtz as a common anti-parasite drug and as a drug for gynecological disorders could be the cause of the high level of resistance in strains of H. pylori.29–32 In addition, in the mini-review article published by Khademi et al3 in Iran, drug resistance data from different strains of H. pylori were collected from different parts of Iran, and the prevalence of Mtz resistance was reported as 61.6%.30
In the literature, limited information exists regarding the mechanism of resistance to Mtz in H. pylori and has not yet been clearly understood. However, some factors that might be relevant to the mechanism of Mtz resistance are as follows: 1) overexpression of TolC homologous genes that upregulates efflux pump activity and reduces activity of nitroreductases and 2) mutational inactivation of the rdxA and/or frxA encoding an oxygen-insensitive NADPH nitroreductase and NAD(P)H flavin oxidoreductase, respectively.32,33 These mutations, without leading to death, could be the cause of drug resistance during the clinical application.34 Therefore, in agreement with previous studies we have confirmed that Mtz should not be the choice for the first line H. pylori eradication therapy.35,36
In the present study, among Mtz-resistant strains, 63 (92.7%) cases possessed mutation in one or both of the rdxA and frxA, and the most common mutations were due to a premature stop codon (substitution of single amino acid) in the rdxA (40.6%) and frxA (24.6%), while 8.2% of Mtz-resistant strains did not contain any alteration in any of the rdxA and frxA. In the study conducted by Abdollahi et al in Iran, among 35 resistant H. pylori isolates only 22.9% (8/35 isolates) deletion mutations were detected in the rdxA, while in the study performed by Teh et al,25 89.1% of Mtz-resistant strains possessed a premature termination codon or insertion/deletion at one or both of the rdxA and frxA.25,37 Therefore, screening for alteration in the rdxA and frxA could be appropriate to identify about 92% of Mtz-resistant strains of H. pylori in the local population.
Resistance to Cla is contributed to the failure of H. pylori eradication in developing countries.30 In the present study, the Cla resistance was detected in 24.2% (38/157) of isolates, and was much lower than the resistance to Mtz. In the mini-review article that was published by Khademi et al,3 the Cla resistance in strains of H. pylori was detected in 22.4% of isolates, while the low resistance to Cla was reported from neighboring Asian countries, including Egypt (4%), Israel (8.2%), Saudi Arabia (4%), and Kuwait (0%).30,38,39 Therefore, the rate of Cla-resistant strains of H. pylori, in Iran is higher than in the Middle East and developed countries. The three most frequent point mutations, the A2143G, A2142G, and A2142C in the 23S rRNA component of ribosomes were related to Cla resistances in H. pylori.40–42 In the present study, the obtained sequences were aligned with the reference strain 26695 of H. pylori, and out of 38 Cla-resistant strains, 37 (97.3%) cases had mutations, such as the A2143G and A2142G. The A2143G (63.1%) was the most common point mutation in the 23S rRNA, followed by 34.2% of the A2142G substitution. There was no point mutation in the sensitive strains of H. pylori. Other mutations that have been observed in Cla-resistant H. pylori isolates were the A2142C and G2141A, G2224A, G2144T, and C2245T. Therefore, Cla-resistant strains could be detected in 97.3% (37/38) of the local population, by screening for mutations in conserved V domain of genes, encoding the 23S rRNA. In the study conducted by Teh et al,25 V domain of the 23S rRNA was sequenced in 14 Cla-resistant strains, and by screening for the A2142G or A2143G mutations of the 23S rRNA, they were able to detect 92.9% of Cla-resistant strains in the Malaysian population.25
The uncontrolled use of antibiotics may have led to the emergence of MDR strains of H. pylori. In the present study, MDR strains were detected in 9.6% (15/157) of H. pylori isolates. The truncated rdxA (caused by premature stop codon) and deletion/insertion were detected as the most common mutational inactivation in the rdxA and frxA. Double point mutations were only detected in two MDR strains, including the ST3204: A2143G, G2224A and ST3188: A2143G, C2245T. Overall, no correlation was found between levels of MIC between Cla resistance and the point mutation in MDR strains.
Our findings suggested that the efflux pump system was overexpressed in the MDR strains of H. pylori, suggesting that the long-term and inappropriate administration of antibiotics in the clinic may be responsible for the appearance of these strains. Chromosomally encoded drug resistances in H. pylori via overexpression of the efflux pump systems were associated with MDR to antibiotics.43–45 In the present study, based on RT-PCR, our findings suggested that the efflux pump hefA were overexpressed in the MDR strains of H. pylori. The relative gene expression of hefA vs gyrB in 80% (12/15) of clinical MDR strains of H. pylori was significantly higher (P<0.05) in comparison to five susceptible strains. In general, antimicrobial susceptibilities of the isolates collected from GC, PUD, and NUD patients were compared, and no significant difference (P>0.05) in antimicrobial resistance was observed among these groups.
In spite of these observations, a major limitation of this study is the genome diversity of H. pylori. The main cause of resistance to Mtz and Cla is point mutations, which could influence efflux pump-related antibiotic resistance patterns. Therefore, it is difficult to distinguish between these two antibiotic resistance mechanisms. In conclusion, H. pylori genome evolves and shows wide geographical divergence. The prevalence of resistance to H. pylori antimicrobial agent is rapidly growing in Iran, and MDR strains have recently been reported. Mutational inactivation and the overexpression of efflux pump are two mechanisms that increase the resistance to H. pylori antimicrobial agents and the rate of MDR strains. In Iran, the mutations of rdxA and frxA in Mtz-resistant strains and the A2143G and A2142G mutations of 23S rRNA in Cla-resistant strains were significant, and screening for these mutations could help to determine the most effective anti-H. pylori drugs and to prevent antibiotic resistance.
Acknowledgments
This work is part of PhD thesis of Hamed Goodarzi and supported by the Health Research Institute, Infectious and Tropical Diseases Research Center, (Grant No. 94142). Special thanks to the Research Affairs, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran, for the financial support. Seyed Jalal Hashemi and Ahmad Farajzadeh Sheikh share first authorship.
Author contributions
All authors contributed toward data analysis, drafting and revising the paper, gave final approval of the version to be published and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Pellicano R, Ribaldone DG, Fagoonee S, Astegiano M, Saracco GM, Mégraud F. A 2016 panorama of Helicobacter pylori infection: key messages for clinicians. Panminerva Med. 2016;58(4):304–317. | ||
Gerrits MM, van Vliet AH, Kuipers EJ, Kusters JG. Helicobacter pylori and antimicrobial resistance: molecular mechanisms and clinical implications. Lancet Infect Dis. 2006;6(11):699–709. | ||
Khademi F, Faghri J, Poursina F, et al. Resistance pattern of Helicobacter pylori strains to clarithromycin, metronidazole, and amoxicillin in Isfahan, Iran. J Res Med Sci. 2013;18(12):1056–1560. | ||
Baglan PH, Bozdayi G, Ozkan M, Ahmed K, Bozdayi AM, Ozden A. Clarithromycin resistance prevalence and iceA gene status in Helicobacter pylori clinical isolates in Turkish patients with duodenal ulcer and functional dyspepsia. J Microbiol. 2006;44(4):409–416. | ||
Yu JD, Chen J, Zy L, Zhang XP. Prevalence of Helicobacter pylori resistant to clarithromycin, metronidazole and amoxicillin isolated from pediatric patients in China. World J Pediatr. 2006;1:49–52. | ||
Boyanova L, Gergova G, Nikolov R, et al. Prevalence and evolution of Helicobacter pylori resistance to 6 antibacterial agents over 12 years and correlation between susceptibility testing methods. Diagn Microbiol Infect Dis. 2008;60(4):409–415. | ||
Wueppenhorst N, Stueger H-P, Kist M, Glocker E. Identification and molecular characterization of triple- and quadruple-resistant Helicobacter pylori clinical isolates in Germany. J Antimicrob Chemother. 2009;63(4):648–653. | ||
Ho SL, Tan EL, Sam CK, Goh KL. Clarithromycin resistance and point mutations in the 23S rRNA gene in Helicobacter pylori isolates from Malaysia. J Dig Dis. 2010;11(2):101–105. | ||
Gerrits MM. Role of the rdxA and frxA genes in oxygen-dependent metronidazole resistance of Helicobacter pylori. J Med Microbiol. 2004;53(11):1123–1128. | ||
Bina JE, Alm RA, Uria-Nickelsen M, Thomas SR, Trust TJ, Hancock RE. Helicobacter pylori uptake and efflux: basis for intrinsic susceptibility to antibiotics in vitro. Antimicrob Agents Chemother. 2000;44(2):248–254. | ||
Borges-Walmsley MI, Walmsley AR. The structure and function of drug pumps. Trends Microbiol. 2001;9(2):71–79. | ||
Putman M, van Veen HW, Konings WN. Molecular properties of bacterial multidrug transporters. Microbiol Mol Biol Rev. 2000;64(4):672–693. | ||
Paulsen IT, Chen J, Nelson KE, Saier MH. Comparative genomics of microbial drug efflux systems. J Mol Microbiol Biotechnol. 2001;3(2):145–150. | ||
Saier MH, Tam R, Reizer A, Reizer J. Two novel families of bacterial membrane proteins concerned with nodulation, cell division and transport. Mol Microbiol. 1994;11(5):841–847. | ||
Baucheron S, Imberechts H, Chaslus-Dancla E, Cloeckaert A. The AcrB multidrug transporter plays a major role in high-level fluoroquinolone resistance in Salmonella enterica serovar typhimurium phage type DT204. Microb Drug Resist. 2002;8(4):281–289. | ||
Sheikh AF, Yadyad MJ, Goodarzi H, et al. CagA and vacA allelic combination of Helicobacter pylori in gastroduodenal disorders. Microb Pathog. 2018;122:144–150. | ||
Nouraie M, Latifi-Navid S, Rezvan H, et al. Childhood hygienic practice and family education status determine the prevalence of Helicobacter pylori infection in Iran. Helicobacter. 2009;14(1):40–46. | ||
Akopyanz N, Bukanov NO, Westblom TU, Berg DE. PCR-based RFLP analysis of DNA sequence diversity in the gastric pathogen Helicobacter pylori. Nucleic Acids Res. 1992;20(23):6221–6225. | ||
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–2729. | ||
Yang L. Incidence and mortality of gastric cancer in China. World J Gastroenterol. 2006;12(1):17–20. | ||
Abadi AT, Taghvaei T, Mobarez AM, Carpenter BM, Merrell DS. Frequency of antibiotic resistance in Helicobacter pylori strains isolated from the northern population of Iran. J Microbiol. 2011;49(6):987–993. | ||
Glupczynski Y, Broutet N, Cantagrel A, et al. Comparison of the E test and agar dilution method for antimicrobial suceptibility testing of Helicobacter pylori. Eur J Clin Microbiol Infect Dis. 2002;21(7):549–552. | ||
Mégraud F, Lehours P. Helicobacter pylori detection and antimicrobial susceptibility testing. Clin Microbiol Rev. 2007;20(2):280–322. | ||
Han F, Liu S, Ho B, Yan Z, Yan X. Alterations in rdxA and frxA genes and their upstream regions in metronidazole-resistant Helicobacter pylori isolates. Res Microbiol. 2007;158(1):38–44. | ||
Teh X, Khosravi Y, Lee WC, et al. Functional and molecular surveillance of Helicobacter pylori antibiotic resistance in Kuala Lumpur. PLoS One. 2014;9(7):7:e101481. | ||
Tamura K, Peterson D, Peterson N, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. | ||
Marquez B. Bacterial efflux systems and efflux pumps inhibitors. Biochimie. 2005;87(12):1137–1147. | ||
Mohammadi M, Doroud D, Mohajerani N, Massarrat S. Helicobacter pylori antibiotic resistance in Iran. World J Gastroenterol. 2005;11(38):6009–6013. | ||
Perri F, Villani MR, Festa V, Quitadamo M, Andriulli A. Predictors of failure of Helicobacter pylori eradication with the standard “Maastricht triple therapy”. Aliment Pharmacol Ther. 2001;15(7):1023–1029. | ||
Khademi F, Poursina F, Hosseini E, Akbari M, Safaei HG, Ghasemian Safaei H. Helicobacter pylori in Iran: a systematic review on the antibiotic resistance. Iran J Basic Med Sci. 2015;18(1):2–7. | ||
Farhana F, Jamaiah I, Rohela M, Nm A-A, Nissapatorn V. A ten year (1999–2008) retrospective study of amoebiasis in University Malaya medical centre (UMMC), Kuala Lumpur, Malaysia. Trop Biomed. 2009;26:262–266. | ||
Binh TT, Shiota S, Nguyen LT, et al. The incidence of primary antibiotic resistance of Helicobacter pylori in Vietnam. J Clin Gastroenterol. 2013;47(3):233–238. | ||
Tankovic J, Lamarque D, Delchier JC, Soussy CJ, Labigne A, Jenks PJ. Frequent association between alteration of the rdxA gene and metronidazole resistance in French and North African isolates of Helicobacter pylori. Antimicrob Agents Chemother. 2000;44(3):608–613. | ||
Tanih NF, Okeleye BI, Naidoo N, et al. Marked susceptibility of South African Helicobacter pylori strains to ciprofloxacin and amoxicillin: clinical implications. S Afr Med J. 2010;100(1):49–52. | ||
Goh KL, Navaratnam P. High Helicobacter pylori resistance to metronidazole but zero or low resistance to clarithromycin, levofloxacin, and other antibiotics in Malaysia. Helicobacter. 2011;16(3):241–245. | ||
Ahmad N, Zakaria WR, Mohamed R. Analysis of antibiotic susceptibility patterns of Helicobacter pylori isolates from Malaysia. Helicobacter. 2011;16(1):47–51. | ||
Abdollahi H, Savari M, Zahedi MJ, Moghadam SD, Hayatbakhsh Abasi M. A study of rdxA gene deletion in metronidazole resistant and sensitive Helicobacter pylori isolates in Kerman, Iran. Jundishapur J Microbiol. 2011;4(2):99–104. | ||
Llanes R, Soria C, Nagashima S, et al. Phenotypic and genetic characterization of antimicrobial profiles of Helicobacter pylori strains in Cuba. J Health Popul Nutr. 2010;28(2):124–129. | ||
Mégraud F. H pylori antibiotic resistance: prevalence, importance, and advances in testing. Gut. 2004;53(9):1374–1384. | ||
Hunt RH, Xiao SD, Megraud F, et al. Helicobacter pylori in developing countries. World Gastroenterology Organisation Global Guideline. J Gastrointestin Liver Dis. 2011;20(3):299–304. | ||
Hwang TJ, Kim N, Kim HB, et al. Change in antibiotic resistance of Helicobacter pylori strains and the effect of A2143G point mutation of 23S rRNA on the eradication of H. pylori in a single center of Korea. J Clin Gastroenterol. 2010;44(8):1–543. | ||
Liu ZQ, Zheng PY, Yang PC. Efflux pump gene hefA of Helicobacter pylori plays an important role in multidrug resistance. World J Gastroenterol. 2008;14(33):5217–5222. | ||
Falsafi T, Ehsani A, Niknam V. The role of active efflux in antibiotic - resistance of clinical isolates of Helicobacter pylori. Indian J Med Microbiol. 2009;27(4):335–340. | ||
van Amsterdam K, Bart A, van der Ende A. A Helicobacter pylori TolC efflux pump confers resistance to metronidazole. Antimicrob Agents Chemother. 2005;49(4):1477–1482. | ||
Kutschke A, de Jonge BL. Compound efflux in Helicobacter pylori. Antimicrob Agents Chemother. 2005;49(7):3009–3010. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.