Back to Journals » OncoTargets and Therapy » Volume 13
Genes Regulated by HPV 16 E6 and High Expression of NFX1-123 in Cervical Cancers
Authors Chintala S , Levan J , Robinson K, Quist K, Katzenellenbogen RA
Received 29 February 2020
Accepted for publication 18 May 2020
Published 26 June 2020 Volume 2020:13 Pages 6143—6156
DOI https://doi.org/10.2147/OTT.S251926
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjay Singh
Sreenivasulu Chintala, Justine Levan, Kristin Robinson, Kevin Quist, Rachel A Katzenellenbogen
Department of Pediatrics, Indiana University School of Medicine, Indianapolis, IN, USA
Correspondence: Rachel A Katzenellenbogen
Indiana University School of Medicine, Herman B. Wells Center for Pediatric Research, 1044 W. Walnut Street, Indianapolis, IN 46202, USA
Tel +1 317 278 0107
Fax +1 317 274 0133
Email [email protected]
Purpose: High-risk human papillomaviruses (HR HPV) cause cervical cancer, and in these cancers, HPV type 16 is the most common HR type. The HR viral oncogenes E6 and E7 partner with cellular proteins to drive cancer and modulate immune pathways; previously, we demonstrated in keratinocytes that HPV 16 E6 and high expression of the endogenous host protein partner NFX1-123 led to the increased expression of multiple genes, including Notch1, secretory leukocyte peptidase inhibitor (SLPI), and retinoic acid early transcript 1G (RAET1G). The present study was conducted to determine if NFX1-123 was highly expressed in cervical cancer and if genes increased by NFX1-123 and 16E6 in keratinocytes were also increased in cervical cancers.
Materials and Methods: The Cancer Genome Atlas (TCGA) database and The Human Protein Atlas database were used to compare relative mRNA and protein gene expression, respectively, in the normal cervix and cervical cancers. Formalin-fixed paraffin-embedded (FFPE) normal cervix and HPV 16 positive cervical cancer samples were analyzed for relative protein expression by immunohistochemical staining. Protein expression of a subset of regulated genes was quantified by Western blot of HPV positive and negative cell lines.
Results: Immunohistochemical staining of HPV 16 positive cervical dysplasias and cancers revealed high NFX1-123, Ki67, and Notch1 expression. NFX1 and NFX1L1 mRNA levels were increased in cervical cancers compared to normal cervix in the TCGA database. Fourteen genes previously identified as upregulated in keratinocytes with 16E6 and overexpressed NFX1-123 also had high mRNA expression and selected genes had high protein expression in cervical cancers and cell lines.
Conclusion: In cervical cancer, NFX1-123 is highly expressed, and 16E6 and NFX1-123 together alter the expression of a wide set of genes. The involvement of these genes in cell proliferation, differentiation, invasion, and metastasis provides further insight into potential ways that HR HPVs promote cancer initiation and maintenance.
Keywords: human papillomavirus, Notch1, SLPI, TCGA database, the human protein atlas
Introduction
High-risk human papillomaviruses (HR HPV) are the causative agent of multiple types of cancer and account for approximately 5% of the total global burden of cancer.1 There are six types of cancers that can be caused by HR HPV: cervical, vulvar, vaginal, penile, and anal, and head and neck cancers of the oropharynx. Nearly 100% of cervical, 40–51% of vulvar, 40–80% of vaginal, 36–80% of penile, 90–93% of anal cancers, and up to 71% of oropharyngeal cancers are caused by HR HPV.2–8 HPV causes the majority of oropharyngeal head and neck cancers in the US, rising above cervical cancer incidence rates, and once a person has an HPV-associated anogenital cancer, their risk of a second anogenital cancer increases, ranging from 1.75 fold to nearly 14 fold.7,9 Thus, HR HPV infection and HPV-associated cancers are a significant global health concern. While effective surgical and ablative therapies exist, treatment options are limited and have high toxicities, leading to rising rates of cancer morbidities.10,12 Understanding the biology and molecular underpinnings of HPV-associated cancers may lead to identification of novel and specific therapeutic targets and effective treatment strategies.
HR HPV infection drives oncogenesis primarily through the actions of its major oncoproteins E6 and E7, which target the p53 pathway and pRb pathway, respectively, to promote aberrant cellular proliferation. However, although HR HPV infection and its oncoproteins are required for malignant progression, they are not sufficient for transformation. Accumulation of additional genetic mutations, dysregulation of gene expression, and disruption of cellular pathways are required for the full development of HPV-associated cancers. Exactly what these changes are and how they contribute to the initiation and maintenance of cancer in the context of an HPV infection have not been fully explored. Profiling the mutational and gene expression landscapes of tumors is therefore a key area of ongoing research in the biology of HPV-associated cancers.
Integrated genomic and molecular characterization studies of cervical cancers have reported mutations in a number of critical regulatory genes.13–16 In addition to identifying mutations in HPV-associated cervical cancers, connecting genetic mutations to changes in gene expression and dysregulated pathway cascades is critical to a comprehensive understanding of these cancers, their development, and their progression.17 Identifying specific genes whose expression and function are dysregulated in cervical cancers may lead to the identification of novel therapeutic targets.
NFX1 (Nuclear Transcription Factor, X-Box Binding 1) is a gene whose product was initially identified as a transcriptional repressor that binds to the conserved X box motif of HLA-DRA as well as to other MHC class II genes.18 NFX1 plays a role in regulating the duration of an inflammatory response by limiting the induction period of MHC class II molecules by IFN-gamma.18 Three alternative splice variants of NFX1 have been identified, two of which are expressed in epithelial cells and have distinct functions.19–24 Previously, our laboratory demonstrated that the E6 oncoprotein of HR HPV type 16 (16E6) partners with the longer splice variant isoform of NFX1, NFX1-123, to deregulate several cellular processes and increase expression of multiple genes in primary human foreskin keratinocytes (HFKs).25 NFX1-123 functions synergistically with 16E6 to post-transcriptionally stabilize the mRNA of hTERT, the catalytic subunit of telomerase, increase Notch1 expression,22,25 augment JNK signaling, and enhance epithelial differentiation.26 The partnership between 16E6 and NFX1-123 is thus multifunctional. The study presented here investigates the expression of a subset of the 16E6 and NFX1-123 upregulated genes in primary cervical cancers. We found nearly 70% of the genes upregulated by HPV16E6/NFX1-123 in HFKs were also significantly overexpressed in cervical cancers. Overexpression of novel genes that had not been previously associated with HPV positive cancers, such as SLPI, CEBPD and RAET1G, represents new avenues for further investigation and potential therapeutic targets.
Materials and Methods
Cell Culture
SiHa, Caski and HeLa cervical cancer cell lines were a kind gift of Dr. Denise Galloway, and they were authenticated by confirmation of HPV gene expression. Cell lines were cultured in DMEM with 10% FBS and penicillin and streptomycin. Primary human foreskin keratinocytes were isolated from neonatal foreskins and cultured in EpiLife media (Thermo Fisher Scientific, Grand Island, NY) as described previously supplemented with calcium chloride (60 μM), human keratinocyte growth supplement, and penicillin-streptomycin.26 Human foreskin keratinocytes were considered not human subjects as per the Indiana University IRB. All cell culture studies were approved by the Indiana University IBC.
Immunohistochemistry and Histologic Analysis
Thirty-one normal cervical and 34 HPV 16 positive cervical carcinoma in situ (CIS n=4) or squamous cell carcinoma samples (noted per specimen pathology reports as moderately differentiated n=4; poorly differentiated n=16; invasive n=10) that were formalin-fixed and paraffin-embedded (FFPE) were obtained from the University of Washington HPV Research Group Specimen Repository. These samples were deidentified and considered not human subjects as per the University of Washington and Seattle Children’s Research Institute IRBs. Sections were stained as previously published23 with these modifications for primary antibodies and conditions: anti-NFX1-123 (rabbit polyclonal, 1:1000 dilution, kind gift from Dr. Ann Roman), anti-Notch1 (rabbit polyclonal, 1:100, Cell Signaling Technology, Danvers, MA, USA), anti-Ki67 (mouse, 1:100, Dako, Santa Clara, USA), and anti-Keratin 1 (rabbit polyclonal, 1:500, Thermo Fisher, Waltham, MA, USA). Antibodies were added and incubated with sections on slides overnight at 4°C in a humidified chamber and then washed. Slides were incubated for 30 min at room temperature with Rabbit SignalStain Boost IHC Detection Reagent (Cell Signaling Technology, Danvers, MA, USA), washed, and then incubated with SignalStain DAB (Cell Signaling Technology, Danvers, MA, USA). Sections were counterstained with hematoxylin and washed. The sections were subsequently dehydrated and mounted. Staining intensity was scored through a review of multiple 20x slide images. Scoring was based on the maximum staining intensity of a specimen: none (no stain), low (yellow), moderate (yellow-brown) or high (dark brown). Analysis of the scoring was reviewed and confirmed by all co-authors.
The Cancer Genome Atlas Data Analysis
Though there are seven different cancers associated with HPV infections, we restricted our analysis for this study to cervical cancers. UCSC Xenabrowser (https://xenabrowser.net) was used to obtain normalized mRNA gene expression data in cancer samples from The Cancer Genome Atlas, TCGA Target GTEx.27 Normal tissues of the cervix were identified, and gene expression data from these tissues were used as a normal control. In the TCGA cervical cancer database, gene expression from 303 primary tumors and 10 normal cervical tissues were quantified and compared. Two metastatic tumors and three ‘normal solid tissues’ from the TCGA database were excluded from the primary tumor analysis. Out of the 25 genes previously identified by our laboratory as upregulated two fold or more by 16E6/NFX1-123,25 19 genes had expression data available for analysis. In addition to these genes, we also determined the expression of NFX1 and NFXL1 in the cervical samples. Of note, this browser did not specify any splice variant isoforms of NFX1, and as such did not delineate the expression of individual isoforms, such as NFX1-91 and NFX1-123, the latter of which is the longer splice variant of the NFX1 gene.
The Human Protein Atlas Data Analysis
Genes that were upregulated by 16E6 and NFX1-123 in HFKs,25 along with NFX1 encoded protein expression, were searched in The Human Protein Atlas database (https://www.proteinatlas.org/).28,29 The rabbit polyclonal anti-NFX1 (HPA073519) antibody utilized in The Human Protein Atlas database was generated using an immunogen sequence from the N terminus (AA 49 to 132), and it is expected to recognize both the isoforms of NFX1 (NFX1-91 and NFX1-123) as this region is common to both. Images were filtered for type of cervical cancer in the Pathology Atlas or for matching normal tissue in the Tissue Atlas. Representative image sections were taken from the high-resolution immunohistochemical staining images available in the database. Scores for the immunohistochemical staining intensity were included in the database, low (L), medium (M), or high (H), and they were specified utilizing the validated scoring system from The Human Protein Atlas database. The * symbol in images denotes high protein expression in glandular, as opposed to epithelial, cells.
Western Blot Analysis
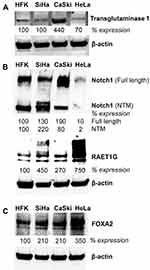
Protein expression of selected genes (RAET1G, Notch1, Transglutaminase1 (TGM1) and FOXA2) were analyzed by Western blot in human foreskin keratinocytes (HFKs), SiHa, CaSki, and HeLa cells. Forty micrograms of whole-cell protein extracts were separated by gel electrophoresis in 4–20% precast Mini-Protein TGX gels and transferred using the Transfer-Blot Turbo transfer pack and the Trans-Blot Turbo Transfer System (Bio-Rad, Hercules, CA, USA). Protein detection was performed with the following primary antibodies: RAET1G (Mouse RAET1G, 1:100 dilution, Santa Cruz Biotechnology, Cat No. sc-53134, Dallas, TX, USA); Notch1 (Rat Notch1 5B5, which detects both full-length Notch1 at 300kDa and its inactive NTM domain at 120kDa, 1:500 dilution, Cell Signaling Technology, Cat No.3447, Danvers, MA, USA); Transglutaminase 1 (Rabbit TGM1 1:500 dilution, Abcam, Cat No. ab27000, Cambridge, MA, USA); FOXA2 (Mouse HNF-3α/β (E-4) FOXA2, 1:100 dilution, Santa Cruz Biotechnology, Cat No. sc-377033, Dallas, TX, USA), and beta-actin (Mouse beta-actin (AC-15), 1:2000 dilution, Invitrogen, Cat No. AM4302, Waltham, MA USA). Membranes were blocked for one hour at room temperature in 2% ECL prime Western blot blocking agent (Amersham GE Healthcare, Pittsburgh, PA, USA), and then primary antibodies were diluted in 2% ECL prime Western blot blocking agent (Amersham GE Healthcare, Pittsburgh, PA, USA) and incubated with membrane blot bound proteins at 4°C overnight. Blots were washed with PBS-T, and then HRP conjugated secondary antibodies were added at a dilution of 1:2000 for RAET1G, TGM1 and FOXA2, or dilution of 1:5000 for Notch1 to 2% ECL prime Western blot blocking agent and incubated at 25 degrees for two hours. Secondary antibody for beta-actin was added at a dilution of 1:10,000 to 2% ECL prime Western blot blocking agent and incubated for one hour. The blots were washed with PBS-T, developed with ECL Prime Western Blot detection reagent, and images were captured with the ChemDoc Imaging unit (Bio-Rad, Hercules, CA, USA).
Image J (NIH, 1.47v, Washington, D.C., USA) quantification of Western blot analyses was used to determine the relative expression of these proteins of interest. For each cell line, band intensity was normalized to its loading control beta-actin, and then fold differences in protein expression were determined relative to the expression level of the protein of interest in HFKs.
Statistical Analysis
Gene expression mRNA data were analyzed for statistical significance using t-test (and nonparametric tests) by choosing Mann–Whitney test and comparison ranks. Results of the Mann–Whitney test were considered significant if the p-value was ≤0.05. All exact p-values are shown (GraphPad Prism 8.0.2 Software, San Diego, CA, USA).
Results
NFX1-123, Proliferation Markers, and Differentiation Markers Were Highly Expressed in Cervical Cancer Specimens
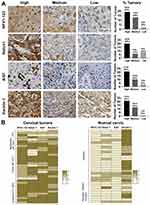
NFX1-123 is a known protein partner of 16E6, and together they upregulate several genes in HFKs that are involved in keratinocyte growth and differentiation including Notch1 and Keratin 1.25 We have previously shown that NFX1-123 itself is upregulated in cervical cancer cell lines24 and primary cervical cancers23 compared to normal cervical tissue, but the full spectrum of NFX1-123 expression across cancers or normal tissue types has not been described. Additionally, the expression of genes, shown to be collaboratively regulated by NFX1-123 and 16E6 in HFKs, has been not examined in the context of cancer. To evaluate the range of expression of NFX1-123 and 16E6/NFX1-123 co-regulated genes, 34 paraffin-embedded formalin fixed HPV 16 positive cervical cancer samples were stained for NFX1-123, Notch1, Ki67, and Keratin1 protein via immunohistochemical staining (IHC). The resulting stained specimens were graded as having high, medium, low or non-detected intensity staining. A representative image of each category of staining and the total number and percentage of tumors in each are presented in Figure 1A. For NFX1-123, 94% (32 out of 34) of tumors had high or medium staining (Figure 1A). High or medium staining was also seen for Notch1, Ki67, and Keratin 1 in 77% (26 out of 34), 91% (31 out of 34), and 97% (33 out of 34) of tumor samples, respectively (Figure 1A). None of these tumors fell within the non-detectable staining category for any of the markers.
To further investigate how the expression of NFX1-123 and these three associated regulated genes shifted in the context of cancer, 31 normal cervical epithelial specimens were stained by IHC for comparison. The staining intensity of both the HPV 16 positive cervical cancers and the normal cervical specimens were then converted to a four-point colorimetric scale, with no detection corresponding to a one and high intensity a four. A heat map for each sample, using this colorimetric scale for all four proteins, is shown in Figure 1B. The cervical samples are clustered by their histologic classification extracted from the HPV Research Group Specimen Repository database (carcinoma in situ, moderately differentiated, poorly differentiated, or invasive squamous cell carcinomas) for cancerous specimens (Figure 1B, left) or by cytologic classification (negative or reactive cellular changes) for normal cervical tissues (Figure 1B, right). Overall, the cervical tumors showed more intense staining than the normal cervical samples for these proteins except for Keratin 1, which was more highly expressed in the normal cervix. Furthermore, IHC analysis of NFX1-123 also revealed high expression (62%,16 out of 26) in the invasive and poorly differentiated tumors as compared to moderately differentiated and CIS samples (38% 3 out of 8) (Figure 2B). In addition to confirming our previous findings that primary cervical tumors have higher overall expression of NFX1-123,23 these data suggest that increased expression of NFX1-123 is more common with worse tumor grades. Finally, these IHC data demonstrate that increased gene expression may also extend to other genes, like Notch1, that are co-regulated by NFX1-123 and 16E6.24
Greater NFX1 and NFXL1 mRNA and NFX1 and NFX1-123 Protein Detected in Human Cervical Cancers
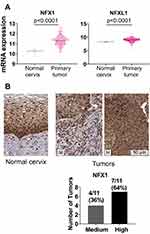
We expanded our analyses of NFX1 and other genes collaboratively regulated by NFX1-123 and 16E6 by using the publicly available databases of TCGA and The Human Protein Atlas. These databases contain an expansive set of cancer types; we chose to focus our investigations on cervical cancer, of which HR HPV is responsible for 99% of cases. In TCGA Target GTEx, we first examined mRNA expression of Nuclear Transcription Factor, X-Box Binding 1 (NFX1) and a gene with NFX1 domain similarity, Nuclear Transcription Factor, X-Box Binding Like 1 (NFXL1),30 comparing their expression in 303 cervical tumors to 10 normal cervical samples. Of note, our previous work has focused specifically on NFX1-123, which is the longer splice variant isoform of NFX1, and the expression status of NFXL1 is not known in HPV-associated cancers. There was a statistically significant upregulation of NFX1 and NFXL1 mRNA in the cervical cancers compared to normal tissue (Figure 2A). Supporting this, 100% of the cervical tumors in the Human Protein Atlas database had medium to high expression of NFX1 (n=11) (Figure 2B). While the protein and the mRNA expression data obtained from these databases did not specify the isoforms of the NFX1 gene that were expressed, these data did corroborate the high expression of NFX1-123 in cervical cancer samples from the biorepository specimens (Figure 1A). Overall, we see high mRNA and protein levels of NFX1 across cervical cancer, suggesting that increased NFX1 expression may have a critical role in HPV-associated cancers.
mRNA Overexpression of 16E6/NFX1-123 Modulated Genes Was a Signature in Cervical Cancer
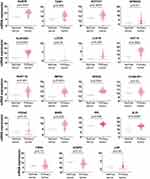
Following our observation that NFX1-123 protein expression, as well as NFX1 mRNA levels, were increased in cervical cancers (Figures 1 and 2), and knowing that 16E6 and NFX1-123 increased a wide range of genes and regulated several cellular pathways in HFKs,24,25,31 we chose to investigate if these same genes were also increased in cervical tumors. We returned to the TCGA Target GTEx database to examine mRNA expression of genes shown to be upregulated twofold or more in HFKs with 16E6 and overexpressed NFX1-123.25 The expression profiles for 19 of the 25 16E6/NFX1-123 upregulated genes were available in TCGA for normal and cancer cervical samples. We found 74% (14 out of 19) of the genes were significantly upregulated in cervical cancers (Figure 3). Five genes, LOR, CEBPD, LCE2B, FOXA2, and FBN2, were deregulated in cervical cancer samples but did not reach statistical significance compared to normal samples due to variability within the population of samples.
Collectively, we found higher expression of NFX1-123, NFX1, and NFXL1 in cervical cancers (Figures 1 and 2), and this high expression was associated with an increase in the majority of genes previously identified as upregulated in 16E6-expressing HFKs with overexpression of NFX1-123.25 To determine if there was a correlation between the relative expression of NFX1 and its associated upregulated genes, we segregated samples into quartiles of NFX1 expression. Then, within the top and bottom quartile of NFX1 expression, we determined the relative expression of the 19 genes in the TCGA database. We did not identify specific relational correlation between high and low NFX1 levels and a matching expression level of any of the 19 genes (data not shown). However, we did note that expression of NFX1 was high across all cervical cancer samples with minimal variation. The lowest and highest fold expression of NFX1 in cervical primary tumors fell between 17.7 and 18.58, making any subset or correlative analysis challenging. Despite this, we do know that these upregulated genes promote proliferation, differentiation and invasion of cancer cells and as such may represent an expression signature for HPV-associated cancers.
Differential Protein Expression of 16E6/NFX1-123 Modulated Genes in Cervical Cancers
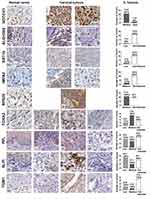
Having seen mRNA increased for 16E6/NFX1-123 regulated genes, we next investigated whether the proteins encoded by these genes were also increased in cervical cancer. A subset of the 19 genes (n=9) available for mRNA expression comparison between normal and tumor tissues were also available for protein analysis in The Human Protein Atlas database. Representative images of the IHC staining for the nine genes are shown for normal cervix (Figure 4, left panels) or cervical tumors (Figure 4, middle panels) as well as quantification of the protein staining (not detected (ND), low, medium, or high) (Figure 4, right panels). In normal tissue, IHC staining was largely non-detected. In tumors, the highest level of staining intensity is described below: 100% (12 of 12) medium to high levels of Notch1; 8% (1 of 12) low levels of ALDH3B2; 17% (2 of 12) low levels of KRT16; 17% (2 of 12) medium levels of IMPA2; 100% (11 of 11) medium levels of RPS29; 75% (9 of 12) medium to low levels of FOXA2; 58% (7 of 12) medium to low levels of PPL; 64% (7 of 11) medium to low levels of SLPI; and 30% (3 of 10) medium to low levels of TGM1.
Therefore, of the nine genes, five had medium to high protein expression in most cervical tumors: PPL, SLPI, FOXA2, RSP29, and Notch1. More specifically, for RSP29 and Notch1, all tumors evaluated had medium to high expression. The IHC staining of HPV 16 positive cervical cancer tissues also revealed high expression of Notch1 (76%, 28 out of 37, Figure 1), further supporting the involvement of Notch1 in cervical cancers. Altogether, these results suggest that PPL, SLPI, and FOXA2, as well as RSP29 and Notch1 may play a critical role in cervical oncogenesis and are potential targets through which 16E6 and NFX1-123 work to promote HPV-associated cancers.
High Protein Expression of 16E6/NFX1-123 Modulated Genes Were Confirmed in HPV Positive Cervical Cancer Cell Lines
Lastly, we conducted expression analyses in three well-characterized HPV positive cervical cancer cell lines to further corroborate the relationship between HPV, NFX1-123, and expression levels of these genes. Western blot analyses of whole-cell protein extracts from SiHa, CaSki (both HPV 16 positive), and HeLa (HPV 18 positive) cell lines were compared to primary human foreskin keratinocytes (HPV negative, HFKs). We have previously shown that NFX1-123 is highly expressed in HPV positive cervical cancer cell lines compared to HFKs.24 Here we demonstrate that RAET1G, Notch1, TGM1, and FOXA2 protein levels were also higher in HPV positive cervical cancer cell lines compared to HFKs (Figure 5). Transglutaminase 1 was highly expressed in CaSki cells. Notch1 had greater protein expression in the two HPV 16 positive cervical cancer cell lines, while RAET1G and FOXA2 were highly expressed in all three cervical cancer cell lines. These results in select genes support the RNA and protein increases seen by immunohistochemical staining and in the TCGA and Protein Atlas databases. Greater NFX1-123 expression and 16E6 co-expression was associated with high expression of several genes that promote growth and differentiation as well as the malignant and metastatic potential of cancer cells.
Discussion
The global incidence of HPV-associated cancers remains high, especially in Asia and sub-Saharan Africa, where HPV is responsible for 30.6% and 15.8%, respectively, of all annual cancer cases.32 Despite the availability of vaccines that protect against infection, insufficient screening and poor vaccination rates limit their efficacy. Furthermore, none of these vaccines cure established infections or diseases; thus, generations of women will be affected by HPV and HPV-associated cancers. Understanding the biology and precise molecular alterations occurring in HPV-associated cancers, and cervical cancer specifically, may lead to identification of new therapeutic strategies and targets.
Previous work in our laboratory identified NFX1-123 as a host protein partner of the E6 protein of HPV type 16.21 A microarray of HFKs expressing 16E6 and overexpressing NFX1-123 revealed a significant upregulation of 25 genes: CCNB1IP1, HSPBL2, TGM1, SLPI, KRT16, SPRR2G, LCE1B, LOC400578, LOC400578, MGC102966, LOC729252, FBN2, RPS29, PPL, RAB7B, CEBPD, ALDH3B2, LCE2B, LOR, BNIPL, RAET1G, LOC643031, FOXA2, IMPA2, and Notch1.25 As a collective, these genes are involved in epithelial differentiation (LCE2B, LCE1B, Notch1, LOR, TGM1); cell growth and survival (PPL, CCNB1IP1, Notch1); inflammation and immune response (SLPI, KRT16, RAET1G, CEBPD); and cellular signaling (FOXA2, CEBPD, IMPA2). Additional studies from our laboratory have confirmed that a set of these genes, and their related pathways, are regulated by NFX1-123 and 16E6 in primary HFKs;25 however, we were also interested in investigating whether these genes were differentially expressed in the greater context of HPV-associated cancers. This study examines the expression of NFX1, NFX1L1, and 16E6/NFX1-123 upregulated genes in HPV-associated primary cervical cancers and normal tissues using The Cancer Genome Atlas (TCGA) database, The Human Protein Atlas database, and immunohistochemical staining of primary cervical samples.
Confirming previous results,23 IHC analysis of cervical biorepository samples revealed higher expression of NFX1-123 in primary cervical cancers compared to normal cervix (Figure 1). Interestingly, a greater percentage of invasive and poorly differentiated tumors had high staining intensity for NFX1-123 (62%, 16 of 26) compared to moderately differentiated tumors and CIS specimens (38%, 3 of 8) (Figure 1). These results suggest not only that greater expression of NFX1-123 may be selected for during the development of cancer, but also that NFX1-123 levels may increase even further over the course of oncogenic progression or in advanced stages of cervical cancer. Analysis of NFX1 and NFXL1 expression in cervical cancers in TCGA and The Human Protein Atlas complemented the IHC staining of cervical biorepository samples; there was increased expression of NFX1 compared to normal tissue (Figure 2).
Although the mechanism that leads to greater expression of NFX1, and NFX1-123, in cervical cancer has not been determined, it was recently demonstrated that NFX1-123 binds to and is a substrate of the deubiquitinase USP9X, whose activity prevents the proteasomal degradation of NFX1-123 protein.33 We also had identified that NFX1-123 bound USP9X, also known as Deubiquitinating Enzyme FAF-X, in a tandem affinity purification assay.21 Keratinocytes immortalized with high-risk HPV E6/E7 showed upregulation of USP9X activity.34 This increase in USP9X activity may be a mechanism by which HR HPV regulates levels of NFX1-123, and through NFX1-123, regulates, in turn, a number of genes that potentially promote cancer initiation and maintenance.
Our analysis of cervical cancers also revealed 14 of 19 genes previously found to be co-regulated in HFKs by 16E6 and NFX1-123 were overexpressed in primary cervical tumors.25 We recognize that further studies will need to be conducted to evaluate the mRNA as well as protein expression of the NFX1-123 isoform in these tumors. These studies will draw relational or functional correlations between the expression levels of NFX1-123 specifically and these genes, rather than associative ones to greater expression of NFX1 as a whole. However, it is reassuring that NFX1 expression and 74% of the co-regulated genes available for analysis were increased in cervical tumors when compared to normal cervical tissue. One important transcript and its encoded protein that was significantly upregulated in cervical cancer is Notch1. Notch1 expression and signaling in cervical cancer are controversial, as its overexpression has been associated with both progression35 and suppression36 of cervical cancer. We have previously seen that in HFKs, 16E6 and NFX1-123 potentiate the expression of Notch1 itself and its canonical and non-canonical signaling pathways,24 although how this role is carried into or changed during cancer development and maintenance has not yet been explored. Notch1 signaling is associated with the activation of the PI3K/Akt pathway and upregulation of c-Myc, two critical oncogenic effectors in cervical cancer,37 and higher expression of Notch1 has been seen in previous studies of cervical cancer development and precancerous lesions.38–40 Notwithstanding, our IHC staining results for Notch1 in cervical biorepository samples demonstrated its greater expression in HPV 16 positive tumor samples (Figure 1). Additionally, our analysis using The Protein Atlas database revealed the higher expression of Notch1 in cervical tumor tissues compared to normal tissue (Figure 4).
We also identified Small Proline-Rich Protein 2G (SPRR2G) as upregulated in cervical cancer in this study; these findings corroborate work from others that SPRR2G is upregulated in HPV-associated vulvar and vaginal cancers.41 Additionally, Cytokeratin 16 (KRT16) was significantly upregulated, and published data have documented upregulation of KRT16 in the cervical cancer cell line, HeLa, compared to HaCaT and HEK293T cells.42
Although their exact functional roles in HPV-associated cancers have not yet been defined, a number of genes in our study have been correlated with other malignancies. Transglutaminase 1 (TGM1) is an enzyme involved in crosslinking of proteins and catenation of polyamines to proteins. It is commonly considered a marker of epithelial differentiation, and its upregulation reflects previous work from our laboratory demonstrating how 16E6 and NFX1-123 mediate epithelial differentiation during active HPV infection.25,26 In the context of cancer, TGM1 functions to promote stemness and chemoresistance in gastric cancer43 and is highly expressed in non-small cell lung cancer cells.44 Fibrillin 2 (FBN2), a large glycoprotein essential for formation of elastic fibers in the extracellular matrix, was reported to be aberrantly methylated in non-small cell lung cancer45 and colorectal cancers.46 The forkhead transcription factor FOXA2, has been shown to coordinate lung cancer tumor growth47 and methylation of its CpG sites were significantly associated with overall survival in oral squamous cell carcinoma.48 We also observed the upregulation of periplankin (PPL) mRNA and protein in cervical cancers. PPL has been reported to be involved in dynamic changes of keratin cytoskeleton.49 Its high expression in esophageal squamous cell carcinomas was correlated with tumor progression, lymph node metastasis, poor prognosis and advanced stage of cancer.50
Our results also revealed the overexpression of genes involved in inflammation and the immune response (SLPI, KRT16, CEBPD, and RAET1G). Secretory leukocyte protease inhibitor (SLPI) promotes cell proliferation, invasion, metastasis, and drug resistance.51,52 In patients with HR HPV positive low or high grade squamous intraepithelial lesions, SLPI concentrations in the cervical mucus were increased compared to controls.53 These results, combined with our finding that SLPI was significantly upregulated in cervical cancers (Figures 3 and 4), merit further investigation of the role of SLPI in cervical cancers. CCAAT/enhancer binding protein delta (CEBPD) functions as transcription factor in cellular differentiation54 and the immune response,55 and its upregulation confers aggressiveness in urothelial carcinoma.56 However, CEBPD expression downregulation was noted in cervical and hepatocellular carcinoma due to epigenetic silencing.57 Retinoic acid early transcript 1G (RAET1G), is a ligand for the NKG2D receptor, mediates natural killer cell cytotoxicity,58 and RAET1G expression was correlated with shorter survival in cervical cancer.59 Again, we found overexpression in cervical cancers and HPV positive cervical cancer cell lines compared to normal tissues and HPV negative cell lines (Figures 3 and 5).
Our analysis of BCL2/adenovirus E1B 19 kD interacting protein like, BNIPL, gene expression at the mRNA level showed significant increase in cervical cancer. However, BNIPL expression was found downregulated in anal cancers (n=13) which is an HPV-associated cancer.60 Furthermore, transcriptome profiling analysis by deep sequencing showed BNIPL downregulation in cervical squamous cancers compared to the normal cervix in patients in China, albeit in a small sample (n=3).61 The differing findings regarding BNIPL levels in HPV-associated cancers may partially be due to the other oncogenic pathways that are altered in cancer cells and regulating BNIPL expression. Functions of IMPA2 in HPV-associated cancers are not known, but in other contexts, IMPA2 is involved in metabolism of carbohydrates, inositols, and phosphatases, and is associated with bone metastasis of breast cancer.62
Poor implementation of global prevention and screening programs mean generations of women remain at risk for cervical and other HPV-associated cancers. Moreover, current treatment options for advanced HPV-associated cancers are suboptimal. It is therefore critical to understand the molecular and cellular dysregulation driven by high-risk HPV in order to identify novel therapeutic targets and strategies for HPV-associated cancers. To elucidate potential targets, we previously utilized HPV 16 transduced human foreskin keratinocytes and identified significant, specific gene expression alterations. Now, these have been confirmed and validated in HPV positive cervical cancers and cell lines. Collectively, these studies highlight novel gene alterations that may serve as therapeutic targets.
Conclusion
In this study, we confirmed the significant increased expression of a subset of genes regulated by HPV 16E6 and its host protein partner NFX1-123, as well as the increase of NFX1-123 itself. Understanding the mechanisms by which 16E6 modulate these genes, the functional consequences upon the tumor cell, and ultimately, the implications for cancer initiation or maintenance will shed the light on potential pathways that may be targeted to effectively eliminate HPV-associated cancers.
Ethics
Deidentified cervical samples were obtained from the University of Washington HPV Research Group Specimen Repository, and they were considered not human subjects as per the University of Washington and Seattle Children’s Research Institute IRBs.
Human foreskin keratinocytes were obtained from neonatal foreskins. They were collected from tissues that would have otherwise been discarded and were anonymous. They were considered not human subjects as per the Indiana University IRB.
SiHa, Caski and HeLa cervical cancer cell lines were a kind gift of Dr. Denise Galloway. The use of these cell lines was approved by the Indiana University IBC.
Acknowledgments
We would like to acknowledge the University of Washington HPV Research Group Specimen Repository for providing cervical cancer and normal cervical tissues. This study was supported by NIH R01 CA 172742 to R.A.K.
Disclosure
All authors report no conflicts of interest in this work.
References
1. Bruni LB-RL, Albero G, Serrano B, et al. Human Papillomavirus and Related Diseases in the World. Summary Report 15 December 2016.: ICO Information Centre on HPV and Cancer (HPV Information Centre); 2016.
2. Daling JR, Madeleine MM, Schwartz SM, et al. A population-based study of squamous cell vaginal cancer: HPV and cofactors. Gynecol Oncol. 2002;84(2):263–270.
3. Daling JR, Madeleine MM, Johnson LG, et al. Penile cancer: importance of circumcision, human papillomavirus and smoking in in situ and invasive disease. IntJCancer. 2005;116(4):606–616.
4. Senkomago V, Henley SJ, Thomas CC, Mix JM, Markowitz LE, Saraiya M. Human papillomavirus-attributable cancers – United States, 2012-2016. MMWR Morb Mortal Wkly Rep. 2019;68(33):724–728. doi:10.15585/mmwr.mm6833a3
5. Tota JE, Chevarie-Davis M, Richardson LA, Devries M, Franco EL. Epidemiology and burden of HPV infection and related diseases: implications for prevention strategies. Prev Med. 2011;53 Suppl 1(S12–21):S12–S21. doi:10.1016/j.ypmed.2011.08.017
6. Chaturvedi AK. Beyond cervical cancer: burden of other HPV-related cancers among men and women. JAdolescHealth. 2010;46(4 Suppl):S20–S26.
7. Chaturvedi AK, Engels EA, Pfeiffer RM, et al. Human papillomavirus and rising oropharyngeal cancer incidence in the United States. J Clin Oncol. 2011;29(32):4294–4301. doi:10.1200/JCO.2011.36.4596
8. Silva Dalla Libera L, Almeida de Carvalho KP, Enocencio Porto Ramos J, et al. Human papillomavirus and anal cancer: prevalence, genotype distribution, and prognosis aspects from midwestern region of Brazil. J Oncol. 2019;2019:6018269.
9. Gilbert DC, Wakeham K, Langley RE, Vale CL. Increased risk of second cancers at sites associated with HPV after a prior HPV-associated malignancy, a systematic review and meta-analysis. Br J Cancer. 2019;120(2):256–268. doi:10.1038/s41416-018-0273-9
10. Cohen PA, Jhingran A, Oaknin A, Denny L. Cervical cancer. Lancet. 2019;393(10167):169–182. doi:10.1016/S0140-6736(18)32470-X
11. Geiger JL, Ku JA. Postoperative treatment of oropharyngeal cancer in the era of human papillomavirus. Curr Treat Options Oncol. 2019;20(3):20. doi:10.1007/s11864-019-0620-y
12. Szturz P, Wouters K, Kiyota N, et al. Low-dose vs. high-dose cisplatin: lessons learned from 59 chemoradiotherapy trials in head and neck cancer. Front Oncol. 2019;9:86. doi:10.3389/fonc.2019.00086
13. Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–582.
14. Cancer Genome Atlas Research N, Albert Einstein College of M, Analytical Biological S, et al.Integrated genomic and molecular characterization of cervical cancer.Nature.2017;543(7645):378–384. doi:10.1038/nature21386
15. Huang J, Qian Z, Gong Y, et al. Comprehensive genomic variation profiling of cervical intraepithelial neoplasia and cervical cancer identifies potential targets for cervical cancer early warning. J Med Genet. 2019;56(3):186–194. doi:10.1136/jmedgenet-2018-105745
16. Perdomo S, Anantharaman D, Foll M, et al. Genomic analysis of head and neck cancer cases from two high incidence regions. PLoS One. 2018;13(1):e0191701. doi:10.1371/journal.pone.0191701
17. Chau NG, Li YY, Jo VY, et al. Incorporation of next-generation sequencing into routine clinical care to direct treatment of head and neck squamous cell carcinoma. Clin Cancer Res. 2016;22(12):2939–2949. doi:10.1158/1078-0432.CCR-15-2314
18. Song Z, Krishna S, Thanos D, Strominger JL, Ono SJ. A novel cysteine-rich sequence-specific DNA-binding protein interacts with the conserved X-box motif of the human major histocompatibility complex class II genes via a repeated Cys-His domain and functions as a transcriptional repressor. J Exp Med. 1994;180(5):1763–1774. doi:10.1084/jem.180.5.1763
19. Gewin L, Myers H, Kiyono T, Galloway DA. Identification of a novel telomerase repressor that interacts with the human papillomavirus type-16 E6/E6-AP complex. Genes Dev. 2004;18(18):2269–2282. doi:10.1101/gad.1214704
20. Xu M, Katzenellenbogen RA, Grandori C, Galloway DA. NFX1 plays a role in human papillomavirus type 16 E6 activation of NFkappaB activity. J Virol. 2010;84(21):11461–11469. doi:10.1128/JVI.00538-10
21. Katzenellenbogen RA, Egelkrout EM, Vliet-Gregg P, Gewin LC, Gafken PR, Galloway DA. NFX1-123 and Poly(A) binding proteins synergistically augment activation of telomerase in human papillomavirus type 16E6 expressing cells. J Virol. 2007;81(8):3786–3796. doi:10.1128/JVI.02007-06
22. Katzenellenbogen RA, Vliet-Gregg P, Xu M, Galloway DA. NFX1-123 increases hTERT expression and telomerase activity posttranscriptionally in human papillomavirus type 16 E6 keratinocytes. J Virol. 2009;83(13):6446–6456. doi:10.1128/JVI.02556-08
23. Vliet-Gregg PA, Robinson KL, Levan J, Matsumoto LR, Katzenellenbogen RA. NFX1-123 is highly expressed in cervical cancer and increases growth and telomerase activity in HPV 16E6 expressing cells. Cancer Lett. 2019;449:106–113. doi:10.1016/j.canlet.2019.02.024
24. Vliet-Gregg PA, Hamilton JR, Katzenellenbogen RA. Human papillomavirus 16E6 and NFX1-123 potentiate Notch signaling and differentiation without activating cellular arrest. Virology. 2015;478:50–60. doi:10.1016/j.virol.2015.02.002
25. Vliet-Gregg PA, Hamilton JR, Katzenellenbogen RA. NFX1-123 and human papillomavirus 16E6 increase Notch expression in keratinocytes. J Virol. 2013;87(24):13741–13750. doi:10.1128/JVI.02582-13
26. Levan J, Vliet-Gregg PA, Robinson KL, Matsumoto LR, Katzenellenbogen RA. HPV type 16 E6 and NFX1-123 augment JNK signaling to mediate keratinocyte differentiation and L1 expression. Virology. 2019;531:171–182. doi:10.1016/j.virol.2019.03.008
27. Goldman M, Craft B, Hastie M, et al. The UCSC Xena platform for public and private cancer genomics data visualization and interpretation. bioRxiv. 2019.
28. Uhlen M, Ponten F. Antibody-based proteomics for human tissue profiling. Mol Cell Proteomics. 2005;4(4):384–393. doi:10.1074/mcp.R500009-MCP200
29. Uhlen M, Zhang C, Lee S, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357(6352):6352. doi:10.1126/science.aan2507
30. Villanueva P, Nudel R, Hoischen A, et al. Exome sequencing in an admixed isolated population indicates NFXL1 variants confer a risk for specific language impairment. PLoS Genet. 2015;11(3):e1004925. doi:10.1371/journal.pgen.1004925
31. Levan J, Vliet-Gregg PA, Robinson KL, Katzenellenbogen RA. Human papillomavirus type 16 E6 and NFX1-123 mislocalize immune signaling proteins and downregulate immune gene expression in keratinocytes. PLoS One. 2017;12(11):e0187514. doi:10.1371/journal.pone.0187514
32. de Martel C, Plummer M, Vignat J, Franceschi S. Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int J Cancer. 2017;141(4):664–670. doi:10.1002/ijc.30716
33. Chen X, Lu D, Gao J, et al. Identification of a USP9X substrate NFX1-123 by SILAC-based quantitative proteomics. J Proteome Res. 2019;18(6):2654–2665. doi:10.1021/acs.jproteome.9b00139
34. Rolen U, Kobzeva V, Gasparjan N, et al. Activity profiling of deubiquitinating enzymes in cervical carcinoma biopsies and cell lines. Mol Carcinog. 2006;45(4):260–269. doi:10.1002/mc.20177
35. Sun Y, Zhang R, Zhou S, Ji Y. Overexpression of Notch1 is associated with the progression of cervical cancer. Oncol Lett. 2015;9(6):2750–2756. doi:10.3892/ol.2015.3143
36. Franko-Tobin LG, Mackey LV, Huang W, et al. Notch1-mediated tumor suppression in cervical cancer with the involvement of SST signaling and its application in enhanced SSTR-targeted therapeutics. Oncologist. 2012;17(2):220–232. doi:10.1634/theoncologist.2011-0269
37. Maliekal TT, Bajaj J, Giri V, Subramanyam D, Krishna S. The role of Notch signaling in human cervical cancer: implications for solid tumors. Oncogene. 2008;27(38):5110–5114. doi:10.1038/onc.2008.224
38. Lathion S, Schaper J, Beard P, Raj K. Notch1 can contribute to viral-induced transformation of primary human keratinocytes. Cancer Res. 2003;63(24):8687–8694.
39. Talora C, Sgroi DC, Crum CP, Dotto GP. Specific down-modulation of Notch1 signaling in cervical cancer cells is required for sustained HPV-E6/E7 expression and late steps of malignant transformation. Genes Dev. 2002;16(17):2252–2263. doi:10.1101/gad.988902
40. Zagouras P, Stifani S, Blaumueller CM, Carcangiu ML, Artavanis-Tsakonas S. Alterations in Notch signaling in neoplastic lesions of the human cervix. Proc Natl Acad Sci U S A. 1995;92(14):6414–6418. doi:10.1073/pnas.92.14.6414
41. Micci F, Teixeira MR, Scheistroen M, Abeler VM, Heim S. Cytogenetic characterization of tumors of the vulva and vagina. Genes Chromosomes Cancer. 2003;38(2):137–148. doi:10.1002/gcc.10263
42. Evstafieva AG, Kovaleva IE, Shoshinova MS, Budanov AV, Chumakov PM. Implication of KRT16, FAM129A and HKDC1 genes as ATF4 regulated components of the integrated stress response. PLoS One. 2018;13(2):e0191107. doi:10.1371/journal.pone.0191107
43. Huang H, Chen Z, Ni X. Tissue transglutaminase-1 promotes stemness and chemoresistance in gastric cancer cells by regulating Wnt/beta-catenin signaling. Exp Biol Med (Maywood). 2017;242(2):194–202. doi:10.1177/1535370216670541
44. Martinet N, Bonnard L, Regnault V, et al. In vivo transglutaminase type 1 expression in normal lung, preinvasive bronchial lesions, and lung cancer. Am J Respir Cell Mol Biol. 2003;28(4):428–435. doi:10.1165/rcmb.2002-0114OC
45. Chen H, Suzuki M, Nakamura Y, et al. Aberrant methylation of FBN2 in human non-small cell lung cancer. Lung Cancer. 2005;50(1):43–49. doi:10.1016/j.lungcan.2005.04.013
46. Hibi K, Mizukami H, Saito M, Kigawa G, Nemoto H, Sanada Y. FBN2 methylation is detected in the serum of colorectal cancer patients with hepatic metastasis. Anticancer Res. 2012;32(10):4371–4374.
47. Camolotto SA, Pattabiraman S, Mosbruger TL, et al. FoxA1 and FoxA2 drive gastric differentiation and suppress squamous identity in NKX2-1-negative lung cancer. Elife. 2018;7.
48. Shen S, Wang G, Shi Q, et al. Seven-CpG-based prognostic signature coupled with gene expression predicts survival of oral squamous cell carcinoma. Clin Epigenetics. 2017;9:88. doi:10.1186/s13148-017-0392-9
49. Long HA, Boczonadi V, McInroy L, Goldberg M, Maatta A. Periplakin-dependent re-organisation of keratin cytoskeleton and loss of collective migration in keratin-8-downregulated epithelial sheets. J Cell Sci. 2006;119(Pt 24):5147–5159. doi:10.1242/jcs.03304
50. Yamada K, Hagiwara T, Inazuka F, et al. Expression of the desmosome-related molecule periplakin is associated with advanced stage and poor prognosis of esophageal squamous cell carcinoma. Transl Cancer Res. 2018;7(1):79–87. doi:10.21037/tcr.2018.01.03
51. Kozin SV, Maimon N, Wang R, et al. Secretory leukocyte protease inhibitor (SLPI) as a potential target for inhibiting metastasis of triple-negative breast cancers. Oncotarget. 2017;8(65):108292–108302. doi:10.18632/oncotarget.22660
52. Munn LL, Garkavtsev I. SLPI: a new target for stopping metastasis. Aging (Albany NY). 2018;10(1):13–14. doi:10.18632/aging.101372
53. Sahin E, Madendag Y, Sahin ME, et al. Cervical local immune response for high-risk human papillomavirus infection: involvement with cervical mucus SLPI proteins. Cancer Control. 2018;25(1):1073274818798598. doi:10.1177/1073274818798598
54. Scott LM, Civin CI, Rorth P, Friedman AD. A novel temporal expression pattern of three C/EBP family members in differentiating myelomonocytic cells. Blood. 1992;80(7):1725–1735. doi:10.1182/blood.V80.7.1725.1725
55. Zannetti C, Bonnay F, Takeshita F, et al. C/EBP{delta} and STAT-1 are required for TLR8 transcriptional activity. J Biol Chem. 2010;285(45):34773–34780. doi:10.1074/jbc.M110.133884
56. Wang YH, Wu WJ, Wang WJ, et al. CEBPD amplification and overexpression in urothelial carcinoma: a driver of tumor metastasis indicating adverse prognosis. Oncotarget. 2015;6(31):31069–31084. doi:10.18632/oncotarget.5209
57. Ko CY, Hsu HC, Shen MR, Chang WC, Wang JM. Epigenetic silencing of CCAAT/enhancer-binding protein delta activity by YY1/polycomb group/DNA methyltransferase complex. J Biol Chem. 2008;283(45):30919–30932. doi:10.1074/jbc.M804029200
58. Bacon L, Eagle RA, Meyer M, Easom N, Young NT, Trowsdale J. Two human ULBP/RAET1 molecules with transmembrane regions are ligands for NKG2D. J Immunol. 2004;173(2):1078–1084. doi:10.4049/jimmunol.173.2.1078
59. Cho H, Chung JY, Kim S, et al. MICA/B and ULBP1 NKG2D ligands are independent predictors of good prognosis in cervical cancer. BMC Cancer. 2014;14(1):957. doi:10.1186/1471-2407-14-957
60. Bruland O, Fluge O, Immervoll H, et al. Gene expression reveals two distinct groups of anal carcinomas with clinical implications. Br J Cancer. 2008;98(7):1264–1273. doi:10.1038/sj.bjc.6604285
61. Lin W, Feng M, Li X, et al. Transcriptome profiling of cancer and normal tissues from cervical squamous cancer patients by deep sequencing. Mol Med Rep. 2017;16(2):2075–2088. doi:10.3892/mmr.2017.6855
62. Smid M, Wang Y, Klijn JG, et al. Genes associated with breast cancer metastatic to bone. J Clin Oncol. 2006;24(15):2261–2267. doi:10.1200/JCO.2005.03.8802
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.