Back to Journals » Cancer Management and Research » Volume 11
Fusobacterium nucleatum prevents apoptosis in colorectal cancer cells via the ANO1 pathway
Authors Lu P, Xu M, Xiong Z, Zhou F , Wang L
Received 29 August 2018
Accepted for publication 10 December 2018
Published 29 October 2019 Volume 2019:11 Pages 9057—9066
DOI https://doi.org/10.2147/CMAR.S185766
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Pei Lu, Minyi Xu, Zhongbo Xiong, Fangfang Zhou, Lei Wang
Department of Clinical Laboratory, Shanghai No. 8 People’s Hospital, Shanghai, China
Correspondence: Pei Lu; Lei Wang
Department of Clinical Laboratory, Shanghai No. 8 People’s Hospital, No. 8, Caobao Road, Xuhui District, Shanghai 200235, China
Tel +86 21 3428 4588; +86 21 3428 4588
Email [email protected];[email protected]
Objective: Chemotherapy failure derived from drug resistance is the most important reason causing the recurrence in colorectal cancer patients. Therefore, it is necessary to shed light on the mechanism of chemotherapy resistance in colorectal cancer patients.
Methods: We looked into the contribution of Fusobacterium nucleatum and ANO1 to chemoresistance in the human colorectal carcinoma cell lines. We silence and overexpress ANO1 in HCT116 and HT29 cells with lentivirus and siRNA knockdown technique in the absence or presence of F. nucleatum, oxaliplatin or 5-fluorouracil (5-FU). ANO1, p-pg, cleaved PARP, cleaved caspase-3, and EGFR expression was measured by Western blot. Cell apoptosis was measured by flow cytometry.
Results: We found that F. nucleatum promoted ANO1 expression on colon cancer cells. Moreover, ANO1 prevent colon cancer apoptosis from oxaliplatin and 5-FU. Additionally, knockdown ANO1 expression could block F. nucleatum protective effects and increase the apoptosis effects induced by oxaliplatin and 5-FU. Therefore, F. nucleatum might be biologically involved in the development of colon cancer chemoresistance via ANO1 pathway.
Conclusions: Taken together, our findings provide a valuable insight into clinical management and therapy, which may ameliorate colorectal cancer patient outcomes.
Keywords: colorectal cancer, F. nucleatum, chemoresistance, 5-fluorouracil, oxaliplatin, ANO1
Introduction
Colorectal cancer (CRC) is the second leading cause of cancer-related death and the third most common cancer in the world.1,2 Chemotherapy drugs can generally shrink tumor size, reduce tumor growth, and inhibit tumor metastasis in advanced CRC patients. For example, 5-fluorouracil (5-FU) and oxaliplatin are commonly used active cytotoxic drugs for CRC patients. 5-FU inhibits the enzyme activity of thymidylate synthase during DNA replication.3 Oxaliplatin inhibits tumor cell growth and causes cell G2 phase arrest by covalently binding DNA and forming platinum-DNA adducts.4 These chemotherapeutic agents were widely combined to treat CRCs1 and are initially effective in most cases. However, patients finally suffer tumor recurrence due to drug resistance, and more than 90% advanced CRC patients die in 5 years.5 Unfortunately, novel immune checkpoint therapy is generally useless for colon cancer patients.6 Therefore, it is necessary to shed light on the mechanism of chemotherapy resistance in CRC patients.
CRC chemoresistance was caused by the complex interplay between the environment and gene regulation. The micropopulation influences tumor-related signaling pathways and intestinal inflammation, which is associated with CRC initiation and progression.7–10 Recent studies have demonstrated that the gut micropopulation may regulate local immune responses and successively impact immunotherapy11,12 and chemotherapy.13,14 Fusobacterium nucleatum (F. nucleatum) is important micropopulation in CRC patients. The amounts of F. nucleatum are gradually augmented from normal tissues to adenoma tissues and to adenocarcinoma tissues in colorectal carcinogenesis.15,16 Furthermore, the abundance of F. nucleatum in CRC tissues is linked to the lower survival rate.17 Especially, compared to those with nonrecurrence post-chemotherapy, CRC patients with recurrence post-chemotherapy contained more F. nucleatum. Another report shows that F. nucleatum can modulate several cellular signal pathways and activate the autophagy pathway which may play a key role in mediating CRC chemoresistance to small drug chemotherapeutics.18
Anoctamin-1 (ANO1) is one of the human chloride channel proteins and is encoded by the ANO1 gene located on 11q13,19 which is frequently amplified in different types of human carcinomas.20,21 The expression of ANO1 is usually upregulated in several cancers including breast cancer,22 prostate cancer,23 gastrointestinal stromal tumor,21,24 colorectal cancer,25,26 esophageal squamous cell carcinoma27 and so on. It also plays an important role in the development of distant metastasis and poor prognosis of cancer patients.23,28,29 Recently, ANO1 has been reported to activate the mitogenactivated protein kinase (MAPK) signaling pathway, which promoted tumorigenesis and invasion.24 Furthermore, ANO1 has been reported to induce the activation of EGFR and calmodulin-dependent protein kinase II and subsequently activate serine-threonine protein kinase (AKT) and MAPK signaling in breast cancer, which contribute to cancer progression.22 Researches from two different groups have demonstrated that high ANO1 expression was a significant prognostic factor for overall survival of patients with CRC and inhibition of ANO1 expression suppresses growth and invasion in human CRC cells.25,26 However, a detailed analysis of the role of ANO1 in CRC and its contribution to chemoresistance is missing. In this study, we demonstrated that F. nucleatum could upregulate ANO1 expression in CRC cells and prevent apoptosis induced by chemotherapy drugs. Through constructing ANO1 overexpression and silencing cell lines, we found that ANO1 plays an important role in the process of chemotherapy drugs which induces CRC cell apoptosis. Our data indicate that F. nucleatum could prevent CRC apoptosis from chemotherapy drugs via the ANO1 pathway.
Materials and methods
Chemicals
5-FU and oxaliplatin were purchased from Sigma-Aldrich Co. (St Louis, MO, USA, F6627, O9512). TRIzol™ was purchased from Invitrogen (Thermo Fisher Scientific, Waltham, MA, USA).
Cell and cell culture
The human colorectal carcinoma cell lines HCT116 and HT29 cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were maintained in a culture medium consisting of RPMI1640 (Thermo Fisher Scientific) supplemented with 10% FBS (lot No. 40K2368, Sigma-Aldrich or lot No. 1248850, Thermo Fisher Scientific), 100 U/mL penicillin and 100 mg/mL streptomycin (Thermo Fisher Scientific) in an atmosphere of 95% air and 5% CO2 at 37°C.
Recombinant lentiviron packaging
The cDNA (CGAAGAAGATGTACCACAT (837–855 nt, NM_018043.5) targeting siRNA site of ANO1 gene, the pervasive disturbing sequence designed as negative-control shRNA and the gene coding ANO1 (NM_018043.5) were cloned into lentiviral nuclear vectors which produced pLKO.1-shANO1, pLKO.1-siNC and pLVX-Puro-ANO1 by JRDUN Biotechnology Co., Ltd (Shanghai, China), respectively. 293 T cells (ATCC) were transfected with the pLKO.1-shANO1, pLKO.1- siNC or pLVX-Puro-ANO1 mixing the lentiviral packaging plasmids (psPAX2 and pMD2G) using lipofectamine RNAiMAX (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. After 6 hours, the culture medium was changed to complete medium. The supernatant was concentrated and collected after culturing for 48 hours. The virus titer of the 293 T cells was determined using the dilution gradient method. The transfected cells were stored in a –80°C refrigerator for later use.
Examination of cell transfection
Three packaged recombinant lentivirus were separately diluted into three groups with culture medium with 5 μg/mL polybrene and were transfected into HCT116 and HT29 cells, respectively. The culture medium was changed to complete medium at 12 hours after transfection. Cells were cultured for 72 hours, following which the expression level of GFP in the cells was observed using an inverted fluorescence microscope. The transfection rate was also analyzed using Image- Pro Plus 6.0 software (Media Cybernetics, Inc., Rockville, MD, USA). The lentivirus-infected cells with the GFP+ were sorted with flow cytometry. Knockdown and overexpression of ANO1 gene in both cell lines were assessed by Western blot and RT-PCR.
Quantitative RT-PCR
Total RNA was extracted from the GFP+ lentivirus-infected cells using TRIzol reagent (Thermo Fisher Scientific) and quantified using a Merinton SMA1000 instrument (Merinton; Ann Arbor, MI, USA). The first strand of cDNA was synthesized from 2.5 μg of total RNA using the iScript cDNA Synthesis kit (Bio-RadLaboratories Inc., Hercules, California, USA) according to the manufacturer’s instructions. The qRT-PCR was performed using the Quantifast SYBR Green PCR kit (Qiagen NV, Venlo, the Netherlands) in an ABI 7500 system. The primers used for PCR of ANO1 were as follows: forward: 5′-AACGGGACCATGCACGGCTT-3′, reverse: 5′-TGTTGTGGTGGTTGCACGGC-3′. The primers were synthesized by Shanghai Sangon Biological Engineering Technology & Services Co., Ltd (Shanghai, China). The reaction cycles for all genes were initiated as follows: 95°C 30 seconds, 95°C 5 seconds, 64°C 34 seconds, 95°C 15 seconds, 60°C 60 seconds. Forty amplification cycles were necessary to achieve exponential amplification. All experiments were repeated three times. Real-time PCR data were analyzed using the 2−ΔΔCT method.3°
Western blot
Cells were cultured in a 6-well plate at a density of 2×105 cells/well and incubated with oxaliplatin (10 μM) and 5-FU (2 μM) for 24 hours. Following treatments, cells were collected. Total protein was extracted from cells lysed by RIPA buffer (Beyotime Institute of Biotechnology, Shanghai, China) and separated by 10% SDS-PAGE before being transferred onto PVDF membranes (Amersham). The PVDF membranes were blocked with 5% BSA in Tris-buffered saline with TWEEN 20 (TBST) at room temperature (RT) for 2 hours and probed using the primary antibodies against ANO1, p-pg, cleaved PARP, cleaved caspase-3, EGFR and GAPDH at 4°C for 1 hour (1:2,000; all from Cell Signaling Technology, Beverly, MA, USA). The detection was performed using a secondary antibody (Sigma-Aldrich Co.) at room temperature for 1 hour. After washing, the bound antibody was detected with immobilization western chemiluminescent HRP substrate (EMD Millipore, Billerica, MA, USA). Then, chemiluminescent emission was captured on Kodak XAR film, and images were analyzed by ImageJ software. The relative expression of interest protein was represented as the grayscale ratio of the protein to GAPDH. All experiments were repeated four times.
Flow cytometry analysis
Cell apoptosis was detected by two-color immunofluorescence staining in the flow cytometric analysis. Cells (2×105 cells/well) were plated into 6-well plate and then were treated with oxaliplatin (10 μM) and 5-FU (2 μM) for 12 hours. The wells added by PBS were used as negative control. Adherent cells were trypsinized without EDTA before two washing steps with PBS. Cells were then incubated with 5 μL of fluorescein isothiocyanate, annexin V (BD Pharmingen, San Jose, CA, USA) and propidium iodide (BD Pharmingen) in 400 μL of 1 × binding buffer for 15 minutes at RT. Data were analyzed by flow cytometry (BD Biosciences, San Jose, CA, USA) equipped with CellQuest Software (BD Biosciences).
Statistical analyses
All the results are expressed as the mean ± SD. Data were analyzed by single factor analysis of variance and the t-test using SPSS 13.0 software (SPSS Inc., Chicago, IL, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
Silencing and overexpression of ANO1 with shRNA lentivirus
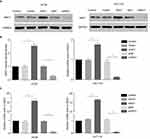
To explore the function of ANO1 in colon cancer cells, HT29 and HCT116 cell lines were separately transfected by lentivirus to silence and overexpress ANO1. Briefly, both the stable silencing of ANO1 and overexpression ANO1 cell lines, which are named siANO1 and ANO1, respectively. Corresponding negative control groups constructed with empty vector lentivirus were named Vector and siNC. The silencing and overexpression efficacy at mRNA and protein levels in colon cancer cell lines were determined by real-time PCR and Western blot, respectively. The results of Western blot showed that the expression levels of ANO1 in the overexpression groups were 1.8–2.5-fold higher than control groups, and the silencing groups ANO1 reduced to 13–28% of that of the control groups (Figure 1A, B). And real-time PCR revealed that the mRNA levels of overexpressed groups were approximately 15.91±1.377 in HT29 and 16.53±0.5425 in HCT116, the silencing groups were approximately 0.1116±0.00761 in HT29 and 0.08067±0.01176 in HCT116 (Figure 1C). The difference in ANO1 mRNA levels between the assay groups and control groups were statistically significant. The results of both Western blot and real-time PCR demonstrated that lentiviral vectors were effective for ANO1 expression.
F. nucleatum promotes ANO1 expression on colon cancer cells
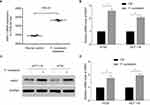
RNA-sequencing data confirmed that ANO1 was expressed in colon cancer cell line HT29, which was in line with most reports while F. nucleatum promoted the ANO1 mRNA levels (Figure 2A, data from GSE90944). We hypothesized that F. nucleatum was biologically involved in the development of colon cancer via ANO1 pathway. To test this hypothesis, we co-cultivated colon cancer cell lines HT29 and HCT116 with F. nucleatum, performed real-time PCR analysis and Western blots analysis, and compared the ANO1 expression profiles between the colon cancer cell lines cocultured with or without F. nucleatum. The rate of ANO1 mRNA expression increased significantly in co-culture groups, and the difference was statistically significant in both cell lines (Figure 2B). Further, protein expression of ANO1 was detected in HT29 and HCT116 cell lines. The rate of ANO1 protein expression was higher in F. nucleatum cocultured cells (Figure 2C, D). Given the fact of ANO1 upregulation in several cancers, and overexpression correlated with the development of distant and poor prognosis of cancer patients, our data suggest that F. nucleatum may cause ANO1 pathway activation and potentially support cancer chemoresistance.
ANO1 prevents colon cancer apoptosis from chemotherapy drugs
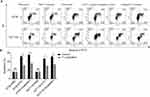
We next hypothesized that F. nucleatum was biologically involved in the development of colon cancer chemoresistance via ANO1 pathway. As expected, oxaliplatin and 5-FU induced HT29 and HCT116 cell apoptosis (Figure 3A, B). Oxaliplatin induced 47.8%±0.8% and 51.6%±0.6% apoptosis in HT29 and HCT116, while 5-FU induced 43.7%±1% and 42.6%±0.9% apoptosis. Co-culture with F. nucleatum reduced HT29 and HCT116 apoptosis induced by these chemotherapeutic agents. Oxaliplatin induced 27.9%±0.2% and 33.1%±0.6% apoptosis in HT29 and HCT116 cocultured with F. nucleatum, while 5-FU induced 26.9%±0.4% and 21.3%±1.5% apoptosis. The apoptosis ratio between CRC cell lines cocultured with or without F. nucleatum was statistically significant. These data indicate that F. nucleatum induces CRC resistance to oxaliplatin and 5-FU.
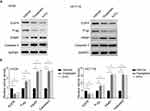
To address whether F. nucleatum prevents CRC apoptosis from chemotherapeutic agents via the ANO1 pathway, we treated ANO1 silencing and overexpression cell lines with chemotherapy drugs. Comparing control groups with F. nucleatum cocultured empty vectors group, F. nucleatum co-culture could reduce the apoptosis ratio. ANO1 silencing could block F. nucleatum protective effects and increase apoptosis ratio. ANO1 overexpression could further reduce the apoptosis ratio and have the lowest apoptosis ratio (Figure 4A, B). Moreover, Western blot data showed that oxaliplatin and 5-FU induced the cleavage of caspase-3 and PARP and downregulation of P-pg and EGFR in HT29 and HCT116 cells (Figure 5A, B). F. nucleatum cocultured ANO1 silencing groups treated with chemotherapy drugs expressed more cleaved caspase-3 and PAPR, while ANO1 overexpression groups expressed less cleaved caspase-3 and PAPR, compared with empty vector group (Figure 6A, B). Meanwhile, F. nucleatum co-culture could induce P-pg and EGFR upregulation. F. nucleatum cocultured ANO1 silencing groups treated with chemotherapy drugs expressed less P-pg and EGFR than ANO1 empty vectors groups. F. nucleatum cocultured ANO1 overexpression groups treated with chemotherapy drugs expressed more P-pg and EGFR than ANO1 empty vector groups. These data suggested the apoptosis effects induced by oxaliplatin and 5-FU could be prevented by ANO1 and F. nucleatum induced ANO1 expression and ANO1 overexpression can further decrease the apoptosis effects. Knockdown ANO1 expression could block F. nucleatum protective effects and increase chemotherapeutic drug-induced apoptosis.
Discussion
CRC is one of the most prevalent carcinomas throughout the world.1,2 CRC patients were usually treated with capecitabine and 5-FU combined with platinum-based chemotherapy4 and initially effective. Unfortunately, drug resistance usually exists and patients will finally die due lack of efficient therapy.2,31 Although novel immune checkpoint therapy can generally cure many other cancer patients,6 it does little to cure patients with CRC and conventional chemotherapy is the first choice of treatment for the latter. Therefore, it is necessary to clarify the mechanisms of chemoresistance in CRC to optimize current treatment strategies. Through a combination of lentivirus and siRNA knockdown technique plus co-culture, we have demonstrated that F. nucleatum was biologically involved in the development of colon cancer chemoresistance via ANO1 pathway.
Cancer epigenetic and modification in CRC chemotherapeutic response have been widely reported.2,32,33 Calcium-activated chloride channel ANO1, also known as transmembrane member 16A (TMEM16A), has been suggested to drive 11q13 amplification by providing growth or metastatic advantage to tumors. ANO1 is rarely expressed in corresponding normal tissues while it is amplified and highly expressed in a large number of cancers. Moreover, transcriptomic and metagenomic examination have showed that F. nucleatum is related to CRC development15,16 and facilitates colorectal carcinogenesis by attaching to the host epithelial E-cadherin via the fusobacterial adhesin FadA34 and fusobacterial lectin Fap2.35 In addition, the amount of F. nucleatum is increased in CRC patients with recurrence postchemotherapy.18 However, it is unknown whether the increasing amount of F. nucleatum and ANO1 are related. In this study, we demonstrated, for the first time, F. nucleatum promotes ANO1 expression on colon cancer cells via co-culture colon cancer cell lines HT29 and HCT116 with F. nucleatum.
Apoptosis is a highly regulated cellular process critical for cell growth and tissue development.36 Loss of apoptosis can result in tumor initiation, growth, and progression.37 5-FU and oxaliplatin can induce tumor cell apoptosis by disrupting and blocking DNA replication.3,4 In agreement, our results showed that oxaliplatin and 5-FU induced HT29 and HCT116 cell apoptosis. Nevertheless, co-culture with F. nucleatum can reduce this effect. Consistently, ANO1 silencing could block F. nucleatum protective effects and increase apoptosis ratio. These facts further demonstrated that F. nucleatum induced ANO1 expression. Several different groups in the world have reported that genetic or pharmacological inhibition of ANO1 can induce apoptosis in different types of cancer cells like gastrointestinal stromal tumor cells,38 human procarcinoma cell27 and so on.39 In line with these findings, we demonstrated that the apoptosis effects induced by oxaliplatin and 5-FU could be prevented by ANO1, F. nucleatum induced ANO1 expression and ANO1 overexpression. Thus, F. nucleatum prevents apoptosis in CRC by the chemicals via the ANO1 pathway. That is to say that F. nucleatum was biologically involved in the development of colon cancer chemoresistance via ANO1 pathway based on the facts that ANO1 is a target of miR-132 that has a crucial role in CRC progression26 and F. nucleatum can inhibit apoptosis via a selective loss of miR-18a* and miR-4802.18 We guess that the putative mechanism by which F. nucleatum prevents apoptosis in CRC by the chemicals via the ANO1 pathway might involve modulation of the amounts of miRNA. ANO1 overexpression induced cell proliferation and inhibition of ANO1 induces apoptosis in prostate carcinoma cells by TNF-α signaling pathway.27 The mechanism is perhaps modulated by cell-specific factors or by the abundance of other anoctamins. Therefore, further investigations are necessary to elucidate the detailed mechanism.
Our work may be helpful for the clinical management of CRC patients besides its biological importance. As F. nucleatum was biologically involved in the development of colon cancer chemoresistance via ANO1 pathway, the conventional chemotherapeutic methods including capecitabine plus oxaliplatin are not appropriate for CRC patients with a high amount of F. nucleatum and anti-F. nucleatum treatment should add to the standard therapy strategies. Further, it also is important to detect F. nucleatum and its associated pathway and to manage patients differentially with different levels of F. nucleatum.
Conclusion
We revealed that F. nucleatum could prevent colon cancer apoptosis from chemotherapy drugs via the ANO1 pathway. Our data suggest that further study is essentially important to reveal the relationship between F. nucleatum, ANO1 and chemoresistance of CRC as well as the potential of F. nucleatum and ANO1 as a therapeutic target of CRC patients.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Cartwright TH. Treatment decisions after diagnosis of metastatic colorectal cancer. Clin Colorectal Cancer. 2012;11(3):155–166.
2. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):63(1):11–30.
3. Walko CM, Lindley C. Capecitabine: a review. Clin Ther. 2005;27(1):23–44.
4. Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7(8):573–584.
5. Dahan L, Sadok A, Formento JL, Seitz JF, Kovacic H. Modulation of cellular redox state underlies antagonism between oxaliplatin and cetuximab in human colorectal cancer cell lines. Br J Pharmacol. 2009;158(2):610–620.
6. Zou W, Wolchok JD, Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: mechanisms, response biomarkers, and combinations. Sci Transl Med. 2016;8(328):328rv4.
7. Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13(11):800–812.
8. Garrett WS. Cancer and the microbiota. Science. 2015;348(6230):80–86.
9. Zitvogel L, Galluzzi L, Viaud S, et al. Cancer and the gut microbiota: an unexpected link. Sci Transl Med. 2015;7(271):271ps1.
10. Man SM, Zhu Q, Zhu L, et al. Critical role for the DNA sensor AIM2 in stem cell proliferation and cancer. Cell. 2015;162(1):45–58.
11. Iida N, Dzutsev A, Stewart CA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342(6161):967–970.
12. Viaud S, Saccheri F, Mignot G, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342(6161):971–976.
13. Sivan A, Corrales L, Hubert N, et al. Commensal bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350(6264):1084–1089.
14. Vétizou M, Pitt JM, Daillère R, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350(6264): 1079–1084.
15. Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22(2):299–306.
16. Kostic AD, Gevers D, Pedamallu CS, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22(2):292–298.
17. Mima K, Nishihara R, Qian ZR, et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut. 2016;65(12):1973–1980.
18. Yu T, Guo F, Yu Y, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017;170(3): 548–563.e16.
19. Katoh M, Katoh M. FLJ10261 gene, located within the CCND1-EMS1 locus on human chromosome 11q13, encodes the eight-transmembrane protein homologous to C12orf3, C11orf25 and FLJ34272 gene products. Int J Oncol. 2003;22(6):1375–1381.
20. Perez-Ordoñez B, Beauchemin M, Jordan RC. Molecular biology of squamous cell carcinoma of the head and neck. J Clin Pathol. 2006;59(5):445–453.
21. Espinosa I, Lee CH, Kim MK, et al. A novel monoclonal antibody against DOG1 is a sensitive and specific marker for gastrointestinal stromal tumors. Am J Surg Pathol. 2008;32(2):210–218.
22. Britschgi A, Bill A, Brinkhaus H, et al. Calcium-activated chloride channel ANO1 promotes breast cancer progression by activating EGFR and CAMK signaling. Proc Natl Acad Sci USA. 2013;110(11):E1026–E1034.
23. Liu W, Lu M, Liu B, Huang Y, Wang K. Inhibition of Ca(2+)-activated Cl(–) channel ANO1/TMEM16A expression suppresses tumor growth and invasiveness in human prostate carcinoma. Cancer Lett. 2012;326(1):41–51.
24. Duvvuri U, Shiwarski DJ, Xiao D, et al. TMEM16A induces MAPK and contributes directly to tumorigenesis and cancer progression. Cancer Res. 2012;72(13):3270–3281.
25. Sui Y, Sun M, Wu F, et al. Inhibition of TMEM16A expression suppresses growth and invasion in human colorectal cancer cells. PLoS One. 2014;9(12):e115443.
26. Mokutani Y, Uemura M, Munakata K, et al. Down-regulation of microRNA-132 is associated with poor prognosis of colorectal cancer. Ann Surg Oncol. 2016;23(Suppl 5):599–608.
27. Song Y, Gao J, Guan L, Chen X, Gao J, Wang K. Inhibition of ANO1/TMEM16A induces apoptosis in human prostate carcinoma cells by activating TNF-α signaling. Cell Death Dis. 2018;9(6):703.
28. Carles A, Millon R, Cromer A, et al. Head and neck squamous cell carcinoma transcriptome analysis by comprehensive validated differential display. Oncogene. 2006;25(12):1821–1831.
29. Carneiro A, Isinger A, Karlsson A, et al. Prognostic impact of arraybased genomic profiles in esophageal squamous cell cancer. BMC Cancer. 2008;8(1):98.
30. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408.
31. Bertotti A, Sassi F. Molecular pathways: sensitivity and resistance to anti-EGFR antibodies. Clin Cancer Res. 2015;21(15):3377–3383.
32. Bardelli A, Siena S. Molecular mechanisms of resistance to cetuximab and panitumumab in colorectal cancer. J Clin Oncol. 2010;28(7): 1254–1261.
33. Dallas NA, Xia L, Fan F, et al. Chemoresistant colorectal cancer cells, the cancer stem cell phenotype, and increased sensitivity to insulin-like growth factor-I receptor inhibition. Cancer Res. 2009;69(5):1951–1957.
34. Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14(2):195–206.
35. Abed J, Emgård JE, Zamir G, et al. Fap2 mediates fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumorexpressed Gal-GalNAc. Cell Host Microbe. 2016;20(2):215–225.
36. Elmore S. Apoptosis: a review of programmed cell death. Toxicol Pathol. 2007;35(4):495–516.
37. Ouyang L, Shi Z, Zhao S, et al. Programmed cell death pathways in cancer: a review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 2012;45(6):487–498.
38. Berglund E, Akcakaya P, Berglund D, et al. Functional role of the Ca2+- activated Cl- channel DOG1/TMEM16A in gastrointestinal stromal tumor cells. Exp Cell Res. 2014;326(2):315–325.
39. Hoffmann EK, Sørensen BH, Sauter DP, Lambert IH. Role of volumeregulated and calcium-activated anion channels in cell volume homeostasis,cancer and drug resistance. Channels (Austin). 2015;9(6):380–396.
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.