Back to Journals » Cancer Management and Research » Volume 12
Functions and Clinical Significance of UPF3a Expression in Human Colorectal Cancer
Authors Bao X, Huang Y, Xu W, Xiong G
Received 2 January 2020
Accepted for publication 19 April 2020
Published 8 June 2020 Volume 2020:12 Pages 4271—4281
DOI https://doi.org/10.2147/CMAR.S244486
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Xinmin Bao,1,* Yuji Huang,2,3,* Weimin Xu,2,3 Gongyou Xiong1
1No.1 People´s Hospital, Jiujiang City, Jiangxi Province, People’s Republic of China; 2Department of Colorectal Surgery, Xin‐Hua Hospital, Shanghai Jiaotong University School of Medicine, Shanghai, People’s Republic of China; 3Shanghai Colorectal Cancer Research Center, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yuji Huang; Gongyou Xiong Email [email protected]; [email protected]
Background: Nonsense-mediated mRNA decay (NMD) can degrade mRNAs with a premature termination codon (PTC), and undegraded mRNAs with PTC mutations can induce a genetic compensation response (GCR) by upregulating its compensatory genes. UPF3a refers to up-frame shift 3A (UPF3a) participating in NMD pathway and GCR. It inhibits the NMD pathway while it stimulates GCR. Notably, the role of UPF3a in cancer remains unclear.
Purpose: The identification and discovery of prognostic markers for colorectal cancer (CRC) are of great clinical significance. The aim of this study was to investigate clinical significance of UPF3a expression in CRC.
Materials and Methods: UPF3a expression was examined in fresh CRC tissues and pared distant metastatic tissues using quantitative real-time PCR, Western blotting and immunohistochemistry staining. Tissue microarray immunohistochemical staining was used to study the relationship of UPF3a with clinicopathological features in 158 CRC patient samples collected from January 2008 to December 2012, and prognosis of CRC was analyzed.
Results: The expression of UPF3a was higher in metastatic tissues than that in primary sites. Moreover, high expression of UPF3a was significantly associated with TNM stage (p=0.009), liver metastasis and recurrence (p< 0.001) in CRC patients. The Cancer Genome Atlas (TCGA) database showed the same trend. In CRC cells, knockdown of UPF3a led to a decline in the migration potential. Kaplan–Meier survival analysis revealed that high UPF3a expression, TNM stage were significantly associated (all P< 0.01) with poor prognosis for patients. Furthermore, univariate and multivariate Cox analysis revealed that high UPF3a expression was independent risk factor for both overall survival and disease-free survival of CRC patients (all P< 0.01).
Conclusion: Results showed that high levels of UPF3a could lead to aggressiveness and poor CRC prognosis. Targeted UPF3a can act as a novel and effective gene therapy for CRC patients to make a better prognosis.
Keywords: UPF3a, colorectal cancer, TCGA, NMD pathway, genetic compensation response, metastasis
Introduction
Colorectal cancer (CRC) is ranked as the world’s fourth most deadly cancer (ranked after lung, liver, and stomach cancer), causing nearly 700,000 deaths annually.1 It has been estimated that 376,300 people were newly diagnosed with CRC, leading to over 191,000 deaths in 2015.2
Notably, metastatic CRC is associated with substantially shorter overall survival (OS) than the early diagnosable nonmetastatic cancer.3 Tumor metastasis can be portrayed as the outcome of a multi-step succession of events, termed as the invasion–metastasis cascade, and has been extensively reviewed previously.4 It is a fundamental biological characteristic of malignant tumors and critically determines cancer prognosis.5 Tumor cell metastasis refers to a process involving tumor cell invasion, circulation, and colonization. Overall, the primary-tumor-driving systemic mechanism, prior to metastasis, can cause tumor cells to be inclined to intravasate into other sites. Currently, the highest incidence rates are observed in developed countries, endangering the lives of numerous men and women. Accordingly, the primary tumor-driving mechanisms and effective predictors associated with cancer progression and metastasis should be examined to help patients monitor and adopt methods for earlier detection and control.
UPF3a is unique among the genes participating in the nonsense-mediated mRNA decay (NMD) pathway and the genetic compensation response (GCR), binding to UPF2, inhibiting the NMD pathway, and recruiting the Wdr-COMPASS complex that induces the GCR. Recently, Ma and Mohamed A have reported that a premature termination codon (PTC)-bearing mRNA could elicit a GCR from Upf3a and COMPASS components in zebrafish,6,7 postulating that NMD participates in GCRs. Furthermore, they demonstrated the role of UPF3a in GCR. In 2006, Shum et al reported that the antagonistic gene paralogs, UPF3a and UPF3b, govern the nonsense-mediated RNA decay,8 demonstrating that UPF3a could inhibit NMD, whereas its paralog, UPF3b, activated NMD. Consistent with its adverse effect on NMD activity and the reduced microsatellite instability (MSI) tumor growth following NMD inhibition, UPF3a expression is noticeably downregulated in MSI CRC versus microsatellite stable (MSS) CRC.9,10 Moreover, Sirkisoon et al have suggested that STAT3 and GLI1/tGLI1, both oncogenic transcription factors, enhance the aggressiveness of triple-negative breast cancers and HER2-enriched breast cancer by upregulating UPF3a expression.11 Remarkably, distant tumor metastases affect patient prognosis.12 Nevertheless, UPF3a participated in both NMD and GCR pathways in tumor; however, the association with patient prognosis in CRC remains poorly investigated.
In the present study, the expression of UPF3a in CRC tissues was detected, and the correlation between UPF3a and the clinicopathological features was analyzed. Importantly, whether UPF3a expression could be a potential prognostic biomarker to predict cancer progression remains of clinical significance.
Materials and Methods
Patients and Samples
A total of 158 CRC samples were harvested in the Department of Colorectal Surgery, Xinhua Hospital, Shanghai Jiaotong University School of Medicine (Shanghai, China), from January 2008 to December 2012. Their formalin-fixed, paraffin-embedded (FFPE) tumor and matched normal mucosa were made into tissue microarray (TMA) for further immunohistochemistry (IHC) analysis.
Cases of fresh paired tissues were collected and analyzed by real-time quantitative polymerase chain reaction (qRT-PCR) and analyzed by Western blots, respectively. The Ethics Committee of Xinhua Hospital Affiliated to Shanghai Jiao Tong University School of Medicine approved this study (No.XHEC-D-2020-057). All patients enrolled in this study have signed the broad consent and the study is strictly in accordance with the Declaration of Helsinki and International Ethical Guidelines for Health-related Research Involving Humans.
Quantitative Real-Time PCR and Western Blotting
Total RNA was extracted and reverse-transcribed with the (TaKaRa, Dalian, China) following the manufacturer’s protocol. To reverse the RNA, the PrimeScript™ RT Master Mix (Takara Biotechnology Co, Ltd.) was used. Then the SYBR Premix ExTaq™ (Takara, Japan) and an Applied Biosystems 7500 Fast Real-Time PCR System (Applied Biosystems, Waltham, MA) were used for qRT-PCR. Relative expression level was normalized to the expression of Actin by the 2−ΔΔCt method.
The sequences of the primers applied are listed as:
UPF3a-F: GCTGTCGGCCCTAGAAGTG UPF3a-R: GAACTCGAAGTAGTCGTGTGC;
WDR5-F: AATTCAGCCCGAATGGAGAGT WDR5-R: AGGCTACATCGGATATTCCCAG;
Actin-F: GCACAGAGCCTCGCCTT Actin-R: GTTGTCGACGACGAGCG.
The UPF3a antibody was purchased from Proteintech Group, Inc. The experiments were performed in triplicate.
Cell Culture
All human CRC cell lines were harvested from the American Type Culture Collection (ATCC). Cells were incubated in Dulbecco’s modified Eagle’s medium (DMEM; Hyclone, Logan, UT, USA) containing 10% fetal bovine serum (Hyclone, Logan, UT, USA) and 1% penicillin/streptomycin, and subsequently maintained at 37 °C in an incubator under a 5% CO2 atmosphere.
Cell Viability Assay
CCK8 method was employed for the cell viability assay. Cells were cultured in 96-well plates (2x103 cells, 100 µL per well). The next day, each well was added with 10 μL CCK8 reagent (Dojindo, Washington, USA) and subsequently incubated at 37 °C for another 1 h. The absorbance was measured at 450 nm with a spectrophotometer.
RNA Interference and Enforced Expression of UPF3a
shRNA sequences targeting UPF3a were cloned in a pLKO.1 vector. With pMD.2G and psPAX2 packaging system, Lentiviruses were generated in 293T cells. Cells received the incubation with supernatant with virus for 2 days, and stable pool cells were taken for 1 week using puromycin.
shUPF3a-1: TACTCAAGAGCATACATTAAT;
shUPF3a-2: GACGTAGAAACACGCAGAAAC;
shUPF3a-3: GATGTGGAGAGATCTCAAGAA.
The coding sequence of UPF3a was cloned by PCR amplification from the complementary DNA of HEK293T cells, which was inserted into a puromycin-resistant lentiviral vector (plvx-Puro) through homologous recombination to construct a plasmid that overexpressed UPF3a, with an empty plvx-Puro vector used as a control.
Transwell and Wound-Healing Assay
To assess the effect of UPF3a knockdown on the cell migration ability, a Transwell assay was first performed. Briefly, 1x105 cells were counted to seed into an FBS-deficient upper chamber, while the lower chamber was added with 500 μL complete medium and incubated for 13 or 48 hr. After washing with PBS, then the upper chamber was fixed with 4% paraformaldehyde for 30 min at room temperature. Finally, 0.1% crystal violet was used to further stain these cells attached with the upper chamber for another 20 min at room temperature. Cell migration ability was assessed according to the number of cells penetrated the upper chamber. In addition, wound-healing assay was also performed to further confirm the effect. A total of 1x106 cells were counted to seed into a six-well plates and cultured with low-serum medium (DMEM containing 1% FBS). When the cell density was 100%, a scratch wound was made in the cell monolayer and the initial image should be immediately obtained. Then the cells were cultured for another 48 hr and the corresponding image was also acquired immediately.
Immunohistochemistry and Evaluation of UPF3a Expression
The TMA were deparaffinized and followed by antigen retrieval with citrate buffer (pH 6.0). Then 3% hydrogen peroxide and 5% goat serum were used to block the endogenous peroxidases and nonspecific antigens, respectively. The primary antibody against UPF3a (1:500, proteintech) were incubated over night at 4°C. The secondary antibody was then applied to the TMA for 1 h at room temperature after washing with phosphate-buffered saline (PBS) three times. Finally, the diaminobenzidine chromogen (Beyotime, Haimen, China) was performed to detect the positive expression of the primary antibody. The TMA ultimately was counterstained with hematoxylin and cover slipped.
After excluding loss of follow-up, 151 eligible patients were ultimately involved in the current research. Immunohistochemistry analysis was conducted following the previous description. The UPF3a expression in TMA was evaluated and semiquantitatively scored based on IHC results by two independent pathologists. The immunohistochemical staining intensity of UPF3a scored as negative (0), weak (1), moderate (2) and strong (3). Besides, the percentage of positive cells scored 5% (0), 5–30% (1), 31–50% (2) and >50% (3). Expression index = % of positive cells × staining intensity. The protein expression in CRC specimens was split into the low expression group (<4) and the high expression group (≥4) for subsequently analysis.
Statistical Analysis
Kaplan–Meier method was performed to assess the survival time distribution, and the Log rank test was used to test significance in DFS and OS among the different prognostic groups. The Cox-proportional hazard model was used to calculate the hazard ratio (HR) for multivariate survival analyses and the confidence intervals (CI) were set at 95%. Data were presented as the values and percentages, or the median and interquartile range (IQR), as appropriate. Unpaired Student’sttest analysis was used for comparison between two different groups. All tests were two‐sided, with p<0.05 considered statistically significant.
Results
UPF3a Was Overexpressed in CRC Cells and Tumor Tissues
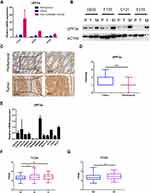
Using real-time, quantitative, reverse-transcription polymerase chain reaction (qRT-PCR), Western blotting, and immunohistochemistry (IHC), the expression of UPF3a in fresh CRC tissues, liver metastatic tissues, and matched peritumoral tissues was detected. As revealed by the qRT-PCR results, UPF3a mRNA was noticeably higher in CRC liver metastatic tissues than in primary tumor tissues (Figure 1A). Simultaneously, Western blotting revealed that the UPF3a protein was more significantly expressed in liver metastatic tissues than in matched primary tumor tissues (Figure 1B). Moreover, UPF3a IHC staining intensity demonstrated a stronger intensity in tumor tissues than in paired peritumoral tissues (Figure 1C). In the stroma, UPF3a expression could result in insignificant differences in the qRT-PCR results between peritumoral and tumor tissues. Based on statistical analysis, the UPF3a intensity in tumor tissues was higher than that in peritumoral tissues (Figure 1D). Furthermore, UPF3a expression was examined in 10 CRC cells and two normal colorectal epithelial cells (CCD841 and NCM460). As shown in Figure 1E, UPF3a expression was elevated in CRC cells compared with that in normal colorectal epithelial cells (Figure 1E). Patients with CRC from The Cancer Genome Atlas (TCGA) database reported an identical trend, revealing that the upregulation of UPF3a correlated with the tumor stage of these patients (Figure 1F), and patients with distant metastasis demonstrated a higher UPF3a expression (Figure 1G).
Alteration of UPF3a Expression Affects Cell Mobility
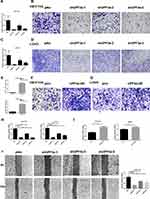
In addition, this study examined the effects of UPF3a knockdown in cancer cells. To this end, stable expression of shRNA in Lovo and HCT116 cells, with three specifically targeted against UPF3a, was achieved. In both cell lines, qRT-PCR and Western blotting assays confirmed shRNA-mediated depletion of UPF3a (Figure 2A, C and Supplementary Fig. A). As shown in Figure 2B and D, the cell migration assay revealed that the UPF3a knockdown impaired Lovo and HCT116 cell mobility. To confirm the effect of UPF3a on the migration ability of CRC cells, we enforced the UPF3a expression in HCT116 cells and LOVO cells. The efficiency of the UPF3a overexpression was confirmed by qRT‐PCR (Figure 2E) and Western blot analysis (Supplementary Fig. B). It was similar to the above results, UPF3a overexpression significantly promoted the cell migration ability compared with the control cells by Transwell assay (Figure 2F and G). Statistical analysis of the results were showed in Figure 2H and I. Based on the Transwell test, an identical result was confirmed using the wound-healing assay in HCT116 cells (Figure 2J).
To explore whether UPF3a depletion affects cell proliferation, the CCK-8 assay was performed in HCT116 cells. Consistent with the previous experiment, UPF3a knockdown also downregulated cell proliferation (Supplementary Fig. C). Wdr5, a component of the COMPASS complex, participates in the GCR interacting with UPF3a. In this study, Wdr5 was downregulated upon UPF3a knockdown (Supplementary Fig. D). Chen et al and Wang et al have observed that Wdr5 promotes cell proliferation and cell mobility in cancer cells. Wdr5 might be a target of UPF3a, which regulates cell functions in CRC; however, how UPF3a affects Wdr5 remains unknown. Future studies should focus on the NMD pathway or the epigenetic regulation of H3K4 methylation.
High UPF3a Expression Correlates with Poor Survival in CRC Patients
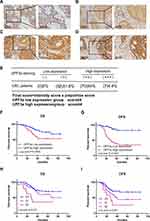
To analyze the association between UPF3a and the clinicopathological features of CRC, the expression of UPF3a in CRC tissue microarray (TMA) was examined using IHC. Correlations analyses suggested that UPF3a expression significantly correlated with the TNM stage (P=0.009) and liver metastasis and recurrence (P<0.001) (Table 1); however, it was not associated with sex, age, or the primary tumor location. As presented in Figure 3A–D, the IHC staining intensities of UPF3a were scored negative (−), weak (+), moderate (++), and strong (+++), and the percentage of positive cells were 5% (−), 5–30% (+), 31−50% (++) and >50% (+++). These scores were calculated by multiplying these two values (0 to 9). In CRC specimens, protein expression was split into low expression (<4) and high expression (>4) for subsequent analysis. As shown in Figure 3E, the low expression group accounted for 51.6% (82), and the high expression group accounted for 48.4% (77). As revealed by Kaplan–Meier analyses, CRC patients demonstrating a higher UPF3a expression reported a poor OS and disease-free survival (DFS) than patients with lower UPF3a expressions (Figure 3F and G). Moreover, tumor stages were confirmed to be associated with OS and DFS (Figure 3H and I). Univariate and multivariate analyses were conducted to determine independent prognostic factors in CRC patients following surgical intervention. Univariate analyses reported that UPF3a expression (P=0.002 for OS and P<0.001 for DFS) and TNM stage (P=0.032 for OS, P=0.027 for DFS) were prognostic factors (Tables 2 and 3), and multivariate analyses indicated that UPF3a expression (P=0.002 for OS, P<0.001 for DFS) was an independent prognostic factor in CRC patients after surgery (Tables 2 and 3).
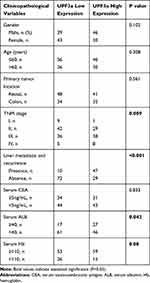 |
Table 1 Correlations of UPF3a Expression with Clinicopathological Characteristics of CRC Patients |
 |
Table 2 Univariable and Multivariable Analysis of OS and Clinicopathological Variables of CRC Patients |
 |
Table 3 Univariable and Multivariable Analysis of DFS and Clinicopathological Variables of CRC Patients |
Discussion
CRC is considered the third most common cancer worldwide, ranking as high as the second leading cause of cancer-related deaths in developed countries.13 The liver is recognized as the most common site of CRC metastasis, as most of the intestinal mesenteric drainage enters the hepatic portal venous system.14 Over 50% of patients with CRC will develop liver metastasis during their lifetime, ultimately resulting in the death of more than two-thirds of these patients.15 Researchers have been committed to improving outcomes, including early detection, effective prognostic indicators of treatment response, and the accurate identification of patients at high risk for recurrence. NMD refers to a highly conserved RNA surveillance pathway that degrades aberrant mRNAs harboring PTCs, approximately 5–20% of the transcripts in a typical transcriptome.6 Early studies have reported that the NMD pathway was initially discovered in Saccharomyces cerevisiae and Caenorhabditis elegans. In recent years, the NMD pathway and its components have been further investigated. Studies performed in yeast to humans have reported that the activation of NMD requires a set of conserved core regulatory factors, the Upf proteins: Upf1, Upf2, and Upf3. Deletion and silencing of each of the genes encoding these factors selectively stabilize PTC-containing transcripts and other NMD substrates.16–19 Mutations in human genes regulating NMD can cause neurodevelopmental disorders, and patients are predisposed to such disorders or have been associated with specific tumor types.20 Furthermore, studies have revealed that the NMD pathway participated in GCR, and interacted with each other.6 Recent studies have demonstrated that the in vivo inhibition of NMD using amlexanox reduces MSI tumor growth, instead of inhibiting MSS tumors.10 Sirkisoon et al have demonstrated that the expression of UPF3a can be elevated by upregulating STAT3 and GLI1/tGLI1, suggesting that UPF3a acts as an oncogene in triple-negative breast cancers and HER2-enriched breast cancer.11 Furthermore, Popp et al have reported that NMD eliminates mutated mRNAs that fail to function in a dominant-negative manner, but are partially functional and could help prevent cancer initiation.21 UPF3a is unique among genes engaging in the NMD pathway and the GCR, possibly playing a balanced role within these two pathways. In this study, UPF3a RNA and protein expression were analyzed in fresh CRC tissues, as well as peritumoral and liver metastatic tissues. To confirm the relationship between UPF3a expression and the prognosis and clinicopathological features of CRC, TMA of CRC tissues were stained for analysis. IHC results revealed that high expression markedly correlated with the TNM stage and metastasis; but it failed to correlate with gender, age, or primary tumor location. The results complied using TCGA data revealed that the UPF3a expression was also higher in patients with poor prognosis stage CRC, and that CRC patients were prone to distant metastasis. In this study, our data reported that the TNM stage and liver metastasis were associated with poor OS and DFS. Importantly, the increased expression of UPF3a was significantly associated with poor OS and DFS in CRC patients, which is consistent with TCGA data. Univariate and multivariate analyses revealed that UPF3a expression and tumor stage were independent prognostic factors in CRC patients after surgery. These results comprehensively demonstrated that the presence of UPF3a closely correlated with poor survival, and UPF3a could act as a novel independent prognostic biomarker in post-surgical CRC patients. However, this study only preliminarily assessed the clinical implications of UPF3a and its function in CRC. Several questions necessitate further investigations, including whether PTC-bearing mRNAs are recruited to the COMPASS complex by Upf3a and guide the complex to upregulate the compensatory genes, and whether PTC-bearing mRNAs activate different types of gene upregulation. Furthermore, studies need to elucidate why some members of gene families are upregulated during GCRs and why only certain genetic knockout mutations do not induce a GCR.
Acknowledgments
This study was supported by grants from the Wu Jieping Medical Foundation (Grant No. 320.6750.19089-33).
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors have declared that no competing interests exist.
References
1. Brody H. Colorectal cancer. Nature. 2015;521:S1. doi:10.1038/521S1a
2. Li J, Qin S, Xu RH, et al. Effect of fruquintinib vs placebo on overall survival in patients with previously treated metastatic colorectal cancer: the FRESCO randomized clinical trial. JAMA. 2018;319(24):2486–2496. doi:10.1001/jama.2018.7855
3. Sun ZQ, Ma S, Zhou QB, et al. Prognostic value of lymph node metastasis in patients with T1-stage colorectal cancer from multiple centers in China. World J Gastroenterol. 2017;23(48):8582–8590. doi:10.3748/wjg.v23.i48.8582
4. Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127(4):679–695. doi:10.1016/j.cell.2006.11.001
5. Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147(2):275–292. doi:10.1016/j.cell.2011.09.024
6. Ma Z, Zhu P, Shi H, et al. PTC-bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components. Nature. 2019;568(7751):259–263.
7. El-Brolosy MA, Kontarakis Z, Rossi A, et al. Genetic compensation triggered by mutant mRNA degradation. Nature. 2019;568(7751):193–197. doi:10.1038/s41586-019-1064-z
8. Shum EY, Jones SH, Shao A, et al. The antagonistic gene paralogs Upf3a and Upf3b govern nonsense-mediated RNA decay. Cell. 2016;165(2):382–395. doi:10.1016/j.cell.2016.02.046
9. El-Bchiri J, Guilloux A, Dartigues P, et al. Nonsense-mediated mRNA decay impacts MSI-driven carcinogenesis and anti-tumor immunity in colorectal cancers. PLoS One. 2008;3(7):e2583. doi:10.1371/journal.pone.0002583
10. Bokhari A, Jonchere V, Lagrange A, et al. Targeting nonsense-mediated mRNA decay in colorectal cancers with microsatellite instability. Oncogenesis. 2018;7(9):70. doi:10.1038/s41389-018-0079-x
11. Sirkisoon SR, Carpenter RL, Rimkus T, et al. Interaction between STAT3 and GLI1/tGLI1 oncogenic transcription factors promotes the aggressiveness of triple-negative breast cancers and HER2-enriched breast cancer. Oncogene. 2018;37(19):2502–2514. doi:10.1038/s41388-018-0132-4
12. Croner RS, Fortsch T, Bruckl WM, et al. Molecular signature for lymphatic metastasis in colorectal carcinomas. Ann Surg. 2008;247(5):803–810. doi:10.1097/SLA.0b013e31816bcd49
13. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–386. doi:10.1002/ijc.29210
14. Lupinacci RM, Paye F, Coelho FF, Kruger JA, Herman P. Lymphatic drainage of the liver and its implications in the management of colorectal cancer liver metastases. Updates Surg. 2014;66(4):239–245. doi:10.1007/s13304-014-0265-0
15. Yeung KT, Yang J. Epithelial-mesenchymal transition in tumor metastasis. Mol Oncol. 2017;11(1):28–39. doi:10.1002/1878-0261.12017
16. Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E. Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J. 2003;22(15):3960–3970. doi:10.1093/emboj/cdg371
17. Cui Y, Hagan KW, Zhang S, Peltz SW. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev. 1995;9(4):423–436. doi:10.1101/gad.9.4.423
18. He F, Brown AH, Jacobson A. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol Cell Biol. 1997;17(3):1580–1594. doi:10.1128/MCB.17.3.1580
19. He F, Jacobson A. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 1995;9(4):437–454. doi:10.1101/gad.9.4.437
20. Jolly LA, Homan CC, Jacob R, Barry S, Gecz J. The UPF3B gene, implicated in intellectual disability, autism, ADHD and childhood onset schizophrenia regulates neural progenitor cell behaviour and neuronal outgrowth. Hum Mol Genet. 2013;22(23):4673–4687. doi:10.1093/hmg/ddt315
21. Popp MW, Maquat LE. Nonsense-mediated mRNA Decay and Cancer. Curr Opin Genet Dev. 2018;48:44–50. doi:10.1016/j.gde.2017.10.007
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.