Back to Journals » OncoTargets and Therapy » Volume 13
FOXK1, Regulated by miR-365-3p, Promotes Cell Growth and EMT Indicates Unfavorable Prognosis in Breast Cancer
Received 18 April 2019
Accepted for publication 27 August 2019
Published 21 January 2020 Volume 2020:13 Pages 623—634
DOI https://doi.org/10.2147/OTT.S212702
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Nicola Silvestris
Fucun Gao, Juan Tian
Breast Department, Linyi Central Hospital, Linyi, Shandong 276400, People’s Republic of China
Correspondence: Juan Tian
Breast Department, Linyi Central Hospital, No. 17 Health Road, Yishui County, Linyi, Shandong 276400, People’s Republic of China
Tel +86 539-2251731 Email [email protected]
Background: Forkhead box K1 (FOXK1) is members of the FOX transcription factor family. Previous work has found out that FOXK1 promotes cell proliferation, migration and invasion in several cancers, such as gastric cancer, glioma cancer and lung cancer; however, the exact role of FOXK1 in breast cancer is still poorly known.
Methods: Here, the association between FOXK1 expression and the clinicopathological characteristics of patients with breast cancer was identified. To further decipher the functional roles of FOXK1, it was overexpressed or knocked down in MCF-7, MDA-MB-231 and MCF-10A cells. Cell Counting Kit-8, colony formation and cell cycle assays were performed to examine the proliferation of breast cancer cells. Moreover, wound-healing and Transwell invasion analyses were carried out to explore the effect of FOXK1 on breast cancer cell migration and invasion.
Results: Our findings discovered that FOXK1 promotes cell proliferation, migration and invasion in breast cancer. In addition, consistent with the previous report, FOXK1 also facilitates EMT in breast cancer. TargetScan was used to predict up-stream of FOXK1, indicating that miR-365-3p could regulate FOXK1 expression in breast cancer.
Conclusion: The findings of the present study demonstrated that miR-365-3p-FOXK1 axis plays a key role in breast cancer.
Keywords: FOXK1, EMT, miR-365-3p, migration, breast cancer
Introduction
Breast cancer has become one of the most common diagnosed cancer among women in the world.1 Metastasis accounts for the majority of morbidity and mortality in breast cancer.2 Although progress has been made in the diagnosis and treatment of breast cancer, the survival rate is still low.3 Therefore, it is an emergency to investigate the underlying molecular mechanism of breast cancer and develop novel therapeutic targets for the treatment.
Forkhead box K (FOXK) family is a subgroup of the Forkhead transcription factors, known to regulate multiple biological processes,4 including cell cycle progression, apoptosis, differentiation, metabolism, survival and senescence.5 FOXK1, also known as MNF, has been found to regulate expression, such as p21, c-myc and cdc2, through recognizing the DNA sequence motif, WRTAAAAYA.6 Previous studies have revealed that FOXK1 induces the epithelial–mesenchymal transition (EMT) in tumour cells, maintains the invasive potential of GC and promotes cell proliferation in ovarian cancer.7,8 These data support a role for human FOXK1 in regulating the developmental process as well as the potential involvement of FOXK1 in tumorigenesis. However, the function and mechanism of FOXK1 in breast cancer remain little known.
EMT is a complicated process necessary during wound healing, embryonic development and tumor progression.9,10 Epithelial cells lose their epithelial features and gain mesenchymal characteristics and become invasive. At the molecular level, the expression of the epithelial marker, such as E-cadherin, is decreased and the expression of mesenchymal markers, such as N-cadherin and vimentin, is increased.11–13 EMT alters the polarity, adhesion ability and improves their migratory and invasive ability.9,10,14 Several transcription factors have been found to be involved in the progress of EMT, such as Slug, Snail and ZEB1.15–18
MicroRNAs (miRNAs) are a class of 22-nucleotide noncoding RNAs encoded by endogenous genes.19 They are known to induce mRNA degradation by binding to the 3′-untranslated region (UTR) of the target.20 Aberrant microRNA expression influences tumor development and progression in breast cancers.21 miR-365 has been found to inhibit breast cancer cell growth and chemo-resistance through GALNT4.22 And miR-365 is considered as a potential diagnostic biomarker for breast cancer.23
In the present study, we examine the function of FOXK1 in the development and progression of breast cancer. We discovered that FOXK1 is up-regulated in breast cancer tissues and cell lines. FOXK1 promotes cell migration and invasion by regulating EMT process in breast cancer. Interestingly, our work found that miR-365-3p is up-stream of FOXK1, so we provided that miR-365-3p-FOXK1 axis plays key roles in the progression of breast cancer.
Materials and Methods
Cell Lines
Human breast cancer MCF-7 cells, MDA-MB-231 and human normal breast epithelial cell line MCF-10A were available from American Type Culture Collection (Manassas, VA, USA). MCF-10A cells were cultured in DMEM/F12 medium supplemented with insulin (10 μg/mL), EGF (10 ng/mL), heat-inactivated horse serum (5%), cholera toxin (1 μg/mL) and hydrocortisone (1 μg/mL). MCF-7 and MDA-MB-231 cells were cultured in DMEM (Fisher Scientific International Inc., Hampton, NH, USA) supplemented with 100 μg/mL penicillin/streptomycin (Fisher Scientific International Inc.) and 10% fetal bovine serum (FBS, Fisher Scientific International Inc.). All cells were cultured at 37°C in a humidified atmosphere with 5% CO2.
Tissue Samples
A total of 93 tissue samples were collected from patients with breast cancer which was diagnosed at the Linyi Central Hospital from March 2005 to April 2013. None of the patients underwent therapy prior to specimen collection, such as chemotherapy and radiotherapy. We had obtained the approval of the ethics committee of the Linyi Central Hospital before this study. Informed consent has been signed by all patients participated in this study.
Western Blotting
Tissue samples and cells were lysed in ice-cold RIPA lysis buffer (Beyotime Institute of Biotechnology) to extract whole protein according to the manufacturer’s protocol. The protein concentration was measured using Pierce™ BCA Protein Assay Kit (Pierce, Waltham, MA, USA). Fifty-microgram protein samples were loaded in each lane, which was further separated by 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose filter membranes (Millipore, Billerica, MA, USA). After blocking in 5% skimmed milk at room temperature for 1 h, the membranes were incubated with indicated antibodies at 4°C overnight and then incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies for 1 h at room temperature. Finally, the blots were detected by enhanced chemiluminescence (Cell Signaling Technology) according to the manufacturer’s protocol. β-Actin was used as an internal control. Antibodies as followed: FOXK1 (no. HPA018864; 1:2000; Sigma-Aldrich, St. Louis, MO, USA), E-cadherin (no. HPA004812; 1:1000; Sigma-Aldrich, St. Louis, MO, USA), N-cadherin (no. HPA058574; 1:1000; Sigma-Aldrich, St. Louis, MO, USA), vimentin (no. HPA001762; 1:2000; Sigma-Aldrich, St. Louis, MO, USA), Slug (no. SAB1305973; 1:2000; Sigma-Aldrich, St. Louis, MO, USA), Snail (no. SAB1306281; 1:2000; Sigma-Aldrich, St. Louis, MO, USA) β-actin (no. A1978; 1:5000; Sigma-Aldrich, St. Louis, MO, USA), Goat Anti-Mouse IgG H&L (HRP) (no. ab205719; 1:3000; Abcam, Cambridge, UK) and Goat Anti-Rabbit IgG H&L (HRP) (no. ab6721; 1:5000; Abcam, Cambridge, UK).
RNA Extraction, Reverse Transcription, and Real-Time PCR
Total RNA was extracted from tissues or cells using TRIzol (Life Technologies) according to the manufacturer’s protocol. Two micrograms RNA was used to synthesize complementary DNA (cDNA) using a poly-A polymerase-based First-Strand Synthesis kit (Takara, Dalian, China). Subsequently, SYBR Green I (Applied Biosystems) was used to perform RT-qPCR. Primers for U6 (no. miRAN0002-1-100) and miR-365-3p (no. MQPS0000357-1-100) were purchased from RiboBio (Guangzhou, China). GAPDH or U6 was used as internal controls for mRNA or miRNA, respective. Relative fold expressions were calculated using 2−∆∆Ct method.24
Cell Counting Kit (CCK-8) Assay
To determine the effect of FOXK1 on cell proliferation, CCK-8 assay was performed according to the manufacturer’s protocol. MCF-7 and MDA-MB-231 cells were transfected with FOXK1 plasmid or FOXK1 siRNA, respectively. After transfection for 24 h, cells were placed into a 96-well plate at a density of 3×103 cells in 200 µL DMEM supplement with 10% FBS. Twenty-microliter CCK-8 was added into each well and cells were incubated for 60 min at 37°C. At 0 h, 24 h, 48h and 72 h, absorbance was measured at 450 nm using an ELx800 microplate reader (BioTek Instruments, Inc., Winooski, VT, USA). Each experiment was carried out three times.
Cell Cycle Analysis
Approximately 4×105 transfected cells were harvested by 0.25% trypsinization and washed with ice-cold phosphate-buffered saline (PBS) three times. After fixation in 75% ice-cold ethanol in PBS at 4°C overnight, cells were washed with ice-cold and incubated with RNase at 37°C at least for 30 min, followed by incubation with propidium iodide (PI) for 20 min at room temperature. Finally, the cell cycle was determined using FACSCanto II flow cytometer (Becton-Dickinson & Co., Franklin Lakes, NJ, USA). Each experiment was carried out three times.
Transwell Invasion Assay
Transwell chamber (BD Biosciences) was used to measure the ability of cells to invade. First, 80 μL Matrigel (Matrigel: DMEM=1:8) was added into Transwell chambers and incubated at 37°C for 30 min to solidify. The upper chamber contained almost 2×104 cells in 200 µL serum-free DMEM. The lower chamber contained 500 μL DMEM with 10% FBS. After incubation at 37°C in an atmosphere containing 5% CO2 for 48 h, cells on the surface of membranes were removed by swab cotton softly and stained with 0.1% crystal violet for 15 min at room temperature. Finally, the invading cells were imaged and counted under a light microscope at ×200 magnification (Olympus Corporation). Each experiment was carried out three times.
Wound-Healing Assay
MDA-MB-231 cells were transfected with FOXK1 plasmid or FOXK1 siRNA for 48 h and placed into 6-well plates at the density of 6×106 cells/well with serum-free DMEM. After scratching using a pipette tip, cells were incubated at 37°C in an atmosphere containing 5% CO2 for 48 h. Images of wound healing were captured under an inverted microscope at ×100 magnification (Olympus Corporation, Tokyo, Japan) at 0 h and 48 h after scratching. Results were analysed with ImageJ software (version 1.2; National Institutes of Health). Each experiment was carried out three times.
Dual-Luciferase Reporter Assay
MCF-7 cells were placed in 12-well plates and co-transfected with miR-365 mimics, renilla and WT-FOXK1 3ʹ-UTR or MT-FOXK1 3ʹ-UTR. After transcription for 24 h, the cells were harvested. The luciferase activity was evaluated using a dual-luciferase reporter assay system (Promega, Madison, WI) according to the manufacturer’s instruction. Firefly luciferase activity was normalized to Renilla luciferase activity. Each experiment was carried out three times.
Statistical Analysis
The results are expressed as the mean ± standard deviation from at least three separate experiments. All data were analyzed using GraphPad Prism 8.0 software (GraphPad Software, Inc., La Jolla, CA, USA). Student’s t-test and one-way analysis of variance followed by the Turkey post hoc test was performed to determine the statistical significance among two groups or multiple groups, respectively. Kaplan–Meier method followed by log-rank test was used to perform survival curves (gepia.cancer-pku.cn) P<0.05 was considered as a statistically significant difference.
Results
FOXK1 Is Up-Regulated in Breast Cancer Tissues and Cells
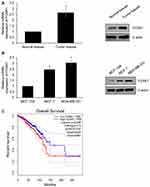
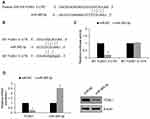
To explore the potential role of FOXK1 in breast cancer, we first detected the FOXK1 mRNA and protein levels in 13 paired primary breast cancer tissues and the corresponding adjacent normal tissues using RT-qPCR and Western blotting analyses. The result revealed that the expression of FOXK1 in primary breast cancer tissues was remarkably higher than those in the adjacent normal breast tissues (Figure 1A). Moreover, we also determine the expression of FOXK1 in breast cancer cell lines, including MCF-7 and MDA-MB-231. Consistent with the expression of FOXK1 in breast cancer tissues, both mRNA and protein levels of FOXK1 were up-regulated in MDA-MB-231 and MCF-7 cells compared with MCF-10A cells (Figure 1B). In addition, as shown in Table 1, we found that the expression of FOXK1 is positively associated with tumor size, TNM stage and lymph node metastasis (Table 1). We also assessed the correlation of FOXK1 and the survival rate using GEPIA website (gepia.cancer-pku.cn), suggesting that high expression of FOXK1 predicted a poor prognosis (Figure 1C). These results suggest that FOXK1 is up-regulated and plays critical roles in breast cancer.
 |
Table 1 Clinic Pathologic Variables in 93 BC Patients |
FOXK1 Promotes the Proliferation of Breast Cancer Cells
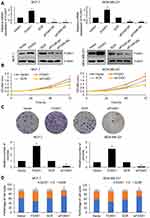
Because of the correlation of FOXK1 expression and tumor size, we assumed that FOXK1 might promote cell proliferation in breast cancer. To verify our hypothesis, we overexpressed or knocked down FOXK1 in MCF-7 and MDA-MB-231 cells, respectively. The expression of FOXK1 was established using RT-qPCR and Western blotting analyses (Figure 2A). To determine the effect of FOXK1 on cell proliferation, we performed CCK-8 assay and colony formation assay. As shown in Figure 2B, ectopic expression of FOXK1 obviously increased the proliferation rate of MCF-7 cells and inhibition of FOXK1 decreased the proliferation rate (Figure 2B). The similar results were found in MDA-MB-231 cells (Figure 2B). The result of colony formation assay revealed that overexpression of FOXK1 resulted in an elevated number of colonies and inhibition of FOXK1 led to a reduced number of colonies (Figure 2C). Next, we further decipher whether FOXK1 promotes cell proliferation through regulation of cell cycle, so we then detected the effect of FOXK1 on cell cycle. We found that FOXK1 could facilitate G1/S phase transition (Figure 2D). Together, our finding indicates that FOXK1 promotes the cell proliferation in breast cancer through facilitating G1/S phase transition.
FOXK1 Improves the Invasive Ability of Breast Cancer Cells
We next determined the potential impact of FOXK1 on breast cancer cell invasion capacity using wound-healing assay and Transwell invasion assay. The number of invaded MDA-MB-231 cells was significantly increased after transfection with ectopic expression-FOXK1 when compared with the mock group (Figure 3A). And inhibition of FOXK1 also promotes the invasion of MDA-MB-231 cells (Figure 3A). Moreover, as shown in Figure 3B, the wound-healing assay indicated that the relative migration distance was increased upon FOXK1 overexpression in MDA-MB-231 cells (Figure 3B). Matrix metalloproteinases (MMPs) are key enzymes for invasion and metastasis, which are the major causes of mortality in breast cancer patients.25 Among them, MMP-2/9 expression is linked with increased metastasis in various tumors, including the brain, prostate, and breast.25,26 So, we then determined the effects of FOXK1 on the expression of MMP-2 and MMP-9 in MDA-MB-231 cells. As shown in Figure 3C, FOXK1 overexpression significantly increased the secretion of MMP-2 and MMP-9, in contrast, FOXK1 inhibition decreased the secretion of MMP-2 and MMP-9 in MDA-MB-231 cells (Figure 3C).
FOXK1 Regulates EMT in Breast Cancer
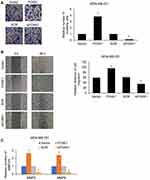
As previously described, FOXK1 increases breast cancer cell migration and invasion capacity. We next explored whether FOXK1 may also be involved in the process of EMT in breast cancer. As shown in Figure 4A, our results demonstrated that ectopic expression of FOXK1 led to increase the mRNA expression of N-cadherin and vimentin; whereas decreased expression of E-cadherin in MCF-10A cells (Figure 4A and B). In contrast, inhibition of FOXK1 in MDA-MB-231 cells significantly decreased the expression of mesenchymal markers N-cadherin and vimentin and improved the expression of epithelial marker E-cadherin (Figure 4A and B). In addition, the expression of EMT-related transcription factors, such as Slug and Snail, was obviously elevated in the FOXK1-overexpressed MCF-10A cells when compared with that of the control cells (Figure 4C and D). In contrast, inhibition of FOXK1 in MDA-MB-231 reduced the expression of Slug and Snail in mRNA and protein levels (Figure 4C and D). Together, our work indicates that FOXK1 regulates EMT in breast cancer.
FOXK1 Is a Target of miR-365-3p in Breast Cancer
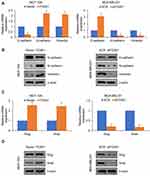
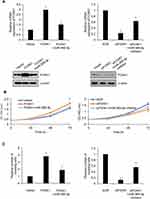
Increasing evidence supports that micro-RNA (miR) plays a key role in human cancer through targeting transcription factors. To further explore the underlying mechanism of FOXK1 in breast cancer, we predicted the up-stream of FOXK1 using TargetScan. The result indicated that FOXF1 might be a target of miR-365-3p (Figure 5A). To further confirm this target association, the dual-luciferase reporter gene plasmid containing the wild-type (WT) or mutant-type MT of FOXK1 3ʹ-UTR (Figure 5B) was constructed. The result of dual-luciferase reporter assays in MCF-7 cells suggested that inhibition of FOXK1 only obviously increased relative luciferase activity of reporter gene with wild-type, but not MT of FOXK1 3ʹ-UTR, suggesting the essential role of miR-365-3p binding site within FOXK1 3ʹ-UTR (Figure 5C). Meanwhile, we also detected the effects of miR-365-3p on the expression of FOXK1. As shown in Figure 5D, we found FOXK1 expression was negatively regulated by miR-365-3p (Figure 5D). To sum up, our finding reveals that FOXK1 is a downstream target of miR-365-3p in breast cancer.
miR-365-3p Reverses the Effects of FOXK1 on Proliferation and Invasion in Breast Cancer Cells
To further investigate whether miR-365-3p was involved in FOXK1-mediated malignant phenotypes of breast cancer cells, MDA-MB-231 cells were co-transfected with pcDNA3.1-FOXK1 plasmids or/and miR-365-3p mimics or co-transfecting siFOXK1 or/and miR-365-3p inhibitor. The mRNA and protein levels of FOXK1 were established using RT-qPCR and Western blotting analyses, respectively (Figure 6A). The results of CCK-8 and Transwell invasion assay demonstrated that up-regulation of miR-365-3p reversed the effects of FOXK1 on proliferation and invasion in MDA-MB-231 cells, and inhibition of miR-365-3p reversed the effects of FOXK1 inhibition on proliferation and invasion in MDA-MB-231 cells, suggesting that miR-365-3p is indeed involved in FOXK1-mediated malignant phenotypes of breast cancer cells (Figure 6B and C).
Discussion
In the present study, FOXK1 expression was up-regulated in breast cancer tissues and cell lines. The expression of FOXK1 was associated with tumor size, TNM stage and metastasis. Moreover, the results of survival curve revealed that high FOXK1 expression predicted a poor prognosis, thus suggesting that FOXK1 may serve as a therapeutic target in breast cancer. In addition, CCK-8, colony formation and cell cycle assays demonstrated that ectopic expression of FOXK1 in breast cancer cells significantly promotes cell proliferation through facilitating G1/S phase transition. Moreover, we found that FOXK1 also reduces cell migration and invasion through the regulation of EMT. EMT is a critical step in cancer metastasis,27,28 epithelial cells change phenotype and obtain the characteristics of mesenchymal cells, gaining the ability to migrate and contribute to tumor metastasis.29 During EMT process, the expression of several proteins is alteration, such as epithelial marker E-cadherin, mesenchymal markers N-cadherin and vimentin, EMT-associated transcription factors TWIST, Snail and Slug.30 Multiple studies have demonstrated that FOXK1 acts as a critical mediator of EMT in certain tumors, such as gastric cancer,31 colorectal cancer32 and prostate cancer,33 indicating that FOXK1 regulates the EMT process and may serve as the core protein during EMT in human cancers.
MicroRNAs (miRNAs) are a class of 22-nucleotide noncoding RNAs encoded by endogenous genes.19 MiRNAs display abnormal expression and functions in multiple kinds of malignancies, which act as different kinds of tumor-related genes.34,35 miR-365-3p has been proven to act as a potential inhibitor in different types of cancer. For example, miR-365-3p inhibits the progression of cutaneous squamous cell carcinoma through regulating cyclin-dependent kinase 6 and 4,36 and miR-365-3p also suppresses tumor growth, proliferation and metastasis in malignant myeloma through targeting NRP.37 Consistent with a previous report, we found that miR-365-3p is down-regulated in breast cancer, and regulates cell proliferation, migration and invasion through targeting FOXK1.
However, there are still present several limitations to our work. First, how FOXK1 regulates proliferation need to be further investigated. RNA-seq and ChIP-seq can perform to reveal the down-stream of FOXK1 in breast cancer. Moreover, whether FOXK1 facilitates tumor proliferation and metastasis needs to be verity in vivo.
In conclusion, our study provides evidence that FOXK1 is up-regulated in breast cancer and serve as an oncogene. Abnormal expression of FOXK1 can promote cell proliferation, migration and invasion. miR-365-3p has been found to down-regulate in breast cancer, here, our work supports that FOXK1 is a target of miR-365-3p, miR-365-3p-FOXK1 axis may as a potential therapeutic strategy for breast cancer.
Disclosure
The authors report no conflicts of interest in this work.
References
1. DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA Cancer J Clin. 2014;64(1):52–62. doi:10.3322/caac.21203
2. Weigelt B, Hu Z, He X, et al. Molecular portraits and 70-gene prognosis signature are preserved throughout the metastatic process of breast cancer. Cancer Res. 2005;65(20):9155–9158. doi:10.1158/0008-5472.CAN-05-2553
3. de la Mare JA, Contu L, Hunter MC, et al. Breast cancer: current developments in molecular approaches to diagnosis and treatment. Recent Pat Anticancer Drug Discov. 2014;9(2):153–175. doi:10.2174/15748928113086660046
4. Kaestner KH, Knochel W, Martinez DE. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000;14(2):142–146.
5. Lam EW, Brosens JJ, Gomes AR, Koo CY. Forkhead box proteins: tuning forks for transcriptional harmony. Nat Rev Cancer. 2013;13(7):482–495. doi:10.1038/nrc3539
6. Wang W, Li X, Lee M, et al. FOXKs promote Wnt/beta-catenin signaling by translocating DVL into the nucleus. Dev Cell. 2015;32(6):707–718. doi:10.1016/j.devcel.2015.01.031
7. Peng Y, Zhang P, Huang X, et al. Direct regulation of FOXK1 by C-jun promotes proliferation, invasion and metastasis in gastric cancer cells. Cell Death Dis. 2016;7(11):e2480. doi:10.1038/cddis.2016.225
8. Li L, Gong M, Zhao Y, Zhao X, Li Q. FOXK1 facilitates cell proliferation through regulating the expression of p21, and promotes metastasis in ovarian cancer. Oncotarget. 2017;8(41):70441–70451. doi:10.18632/oncotarget.19713
9. Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–1428. doi:10.1172/JCI39104
10. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. doi:10.1038/nrm3758
11. Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2(6):442–454. doi:10.1038/nrc822
12. Ferrarotto R, Goonatilake R, Yoo SY, et al. Epithelial-mesenchymal transition predicts polo-like kinase 1 inhibitor-mediated apoptosis in non-small cell lung cancer. Clin Cancer Res. 2016;22(7):1674–1686. doi:10.1158/1078-0432.CCR-14-2890
13. Matsubara D, Kishaba Y, Yoshimoto T, et al. Immunohistochemical analysis of the expression of E-cadherin and ZEB1 in non-small cell lung cancer. Pathol Int. 2014;64(11):560–568. doi:10.1111/pin.12214
14. Shibue T, Weinberg RA. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017;14(10):611–629. doi:10.1038/nrclinonc.2017.44
15. Merikallio H, Turpeenniemi-Hujanen T, Paakko P, et al. Snail promotes an invasive phenotype in lung carcinoma. Respir Res. 2012;13:104. doi:10.1186/1465-9921-13-104
16. Liu CW, Li CH, Peng YJ, et al. Snail regulates Nanog status during the epithelial-mesenchymal transition via the Smad1/Akt/GSK3beta signaling pathway in non-small-cell lung cancer. Oncotarget. 2014;5(11):3880–3894. doi:10.18632/oncotarget.2006
17. Wang M, Zhang R, Zhang S, Xu R, Yang Q. MicroRNA-574-3p regulates epithelial mesenchymal transition and cisplatin resistance via targeting ZEB1 in human gastric carcinoma cells. Gene. 2019;700:110–119. doi:10.1016/j.gene.2019.03.043
18. Grzegrzolka J, Biala M, Wojtyra P, et al. Expression of EMT markers SLUG and TWIST in breast cancer. Anticancer Res. 2015;35(7):3961–3968.
19. Li X, Liu F, Lin B, et al. miR150 inhibits proliferation and tumorigenicity via retarding G1/S phase transition in nasopharyngeal carcinoma. Int J Oncol. 2017;50(4):1097–108.
20. Horman SR, Janas MM, Litterst C, et al. Akt-mediated phosphorylation of argonaute 2 downregulates cleavage and upregulates translational repression of MicroRNA targets. Mol Cell. 2013;50(3):356–367. doi:10.1016/j.molcel.2013.03.015
21. Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi:10.1038/nature03702
22. Zhang J, Zhang Z, Wang Q, Xing XJ, Zhao Y. Overexpression of microRNA-365 inhibits breast cancer cell growth and chemo-resistance through GALNT4. Eur Rev Med Pharmacol Sci. 2016;20(22):4710–4718.
23. Han JG, Jiang YD, Zhang CH, et al. A novel panel of serum miR-21/miR-155/miR-365 as a potential diagnostic biomarker for breast cancer. Ann Surg Treat Res. 2017;92(2):55–66. doi:10.4174/astr.2017.92.2.55
24. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
25. Wang L, Ling Y, Chen Y, et al. Flavonoid baicalein suppresses adhesion, migration and invasion of MDA-MB-231 human breast cancer cells. Cancer Lett. 2010;297(1):42–48. doi:10.1016/j.canlet.2010.04.022
26. Yao J, Xiong S, Klos K, et al. Multiple signaling pathways involved in activation of matrix metalloproteinase-9 (MMP-9) by heregulin-beta1 in human breast cancer cells. Oncogene. 2001;20(56):8066–8074. doi:10.1038/sj.onc.1204944
27. George JT, Jolly MK, Xu S, Somarelli JA, Levine H. Survival outcomes in cancer patients predicted by a partial EMT gene expression scoring metric. Cancer Res. 2017;77(22):6415–6428. doi:10.1158/0008-5472.CAN-16-3521
28. El-Sayed IY, Daher A, Destouches D, et al. Extracellular vesicles released by mesenchymal-like prostate carcinoma cells modulate EMT state of recipient epithelial-like carcinoma cells through regulation of AR signaling. Cancer Lett. 2017;410:100–111. doi:10.1016/j.canlet.2017.09.010
29. Skrypek N, Goossens S, De Smedt E, Vandamme N, Berx G. Epithelial-to-mesenchymal transition: epigenetic reprogramming driving cellular plasticity. Trends Genet. 2017;33(12):943–959. doi:10.1016/j.tig.2017.08.004
30. Choi IJ, Lee NR, Kim SG, et al. Short-term outcomes of endoscopic submucosal dissection in patients with early gastric cancer: a prospective multicenter cohort study. Gut Liver. 2016;10(5):739–748. doi:10.5009/gnl15466
31. Zhang P, Tang WM, Zhang H, et al. MiR-646 inhibited cell proliferation and EMT-induced metastasis by targeting FOXK1 in gastric cancer. Br J Cancer. 2017;117(4):525–534. doi:10.1038/bjc.2017.181
32. Wu M, Wang J, Tang W, et al. FOXK1 interaction with FHL2 promotes proliferation, invasion and metastasis in colorectal cancer. Oncogenesis. 2016;5(11):e271. doi:10.1038/oncsis.2016.68
33. Chen F, Xiong W, Dou K, Ran Q. Knockdown of FOXK1 suppresses proliferation, migration, and invasion in prostate cancer cells. Oncol Res. 2017;25(8):1261–1267. doi:10.3727/096504017X14871164924588
34. Mei Q, Li F, Quan H, Liu Y, Xu H. Busulfan inhibits growth of human osteosarcoma through miR-200 family microRNAs in vitro and in vivo. Cancer Sci. 2014;105(7):755–762. doi:10.1111/cas.12436
35. Song W, Li Q, Wang L, Wang L. Modulation of FoxO1 expression by miR-21 to promote growth of pancreatic ductal adenocarcinoma. Cell Physiol Biochem. 2015;35(1):184–190. doi:10.1159/000369686
36. Zhou L, Wang Y, Ou C, et al. microRNA-365-targeted nuclear factor I/B transcriptionally represses cyclin-dependent kinase 6 and 4 to inhibit the progression of cutaneous squamous cell carcinoma. Int J Biochem Cell Biol. 2015;65:182–191. doi:10.1016/j.biocel.2015.06.009
37. Bai J, Zhang Z, Li X, Liu H. MicroRNA-365 inhibits growth, invasion and metastasis of malignant melanoma by targeting NRP1 expression. Cancer Biomark. 2015;15(5):599–608. doi:10.3233/CBM-150500
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.