Back to Journals » OncoTargets and Therapy » Volume 13
FKBP10 Acts as a New Biomarker for Prognosis and Lymph Node Metastasis of Gastric Cancer by Bioinformatics Analysis and in Vitro Experiments
Authors Gong LB, Zhang C, Yu RX, Li C, Fan YB, Liu YP , Qu XJ
Received 9 March 2020
Accepted for publication 19 June 2020
Published 28 July 2020 Volume 2020:13 Pages 7399—7409
DOI https://doi.org/10.2147/OTT.S253154
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Leo Jen-Liang Su
Li-Bao Gong,1,2,* Chuang Zhang,1,2,* Ruo-Xi Yu,1,2 Ce Li,1,2 Yi-Bo Fan,1,2 Yun-Peng Liu,1,2 Xiu-Juan Qu1,2
1Department of Medical Oncology, The First Hospital of China Medical University, Shenyang 110001, People’s Republic of China; 2Key Laboratory of Anticancer Drugs and Biotherapy of Liaoning Province, The First Hospital of China Medical University, Shenyang 110001, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xiu-Juan Qu
Department of Medical Oncology, The First Hospital of China Medical University, Shenyang 110001, People’s Republic of China
Tel +86-24-83282312
Fax +86-24-83282543
Email [email protected]
Purpose: To explore the role of FKBP prolyl isomerase 10 (FKBP10) protein in the progression of gastric cancer.
Methods: Four independent gastric cancer databases (GSE27342, GSE29272, GSE54129 and TCGA-STAD) were used to identify differentially expressed genes (DEGs). Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was used to identify the abnormally active pathways in patients with gastric cancer. Univariate Cox regression analysis was used to identify genes with stable prognostic value in gastric cancer patients based on three independent gastric cancer databases (GSE15459, GSE62254, TCGA-STAD). Gene set enrichment analysis (GSEA) was used to explore the possible pathways related to FKBP10. The reverse transcription-polymerase chain reaction (RT-PCR) was employed to determine the expression of FKBP10 mRNA in the HGC-27 and MKN-7 cell lines. Adhesion assay was used to detect changes in cell adhesion ability. FKBP10, ITGA1, ITGA2, ITGA5, ITGAV, ITGA6, P-AKT473, P-AKT308, AKT, and β-actin were evaluated by Western blot (WB).
Results: We first performed differential expression genes (DEGs) screening of four independent GC databases (GSE27342, GSE29272, GSE54129 and TCGA-STAD). Eighty-nine genes showed consistent up-regulation in GC, the results of pathway analysis showed that they were related to “Focal adhesion”. The prognostic value of these 89 genes was tested in three independent GC databases GSE15459, GSE62254 and TCGA-STAD cohort. Finally, 12 genes, in which the expression of FKBP10 was prominently increased in patients with lymph node metastasis (LNM), showed stable prognostic value. The following gene set enrichment analysis (GSEA) also showed that FKBP10 is mainly involved in cell adhesion process, while adhesion experiments confirmed that cell adhesion was down-regulated after silencing FKBP10 in GC cells, and adhesion-related molecules integrin αV and α 6 were down-regulated.
Conclusion: FKBP10 may be used as a marker for lymph node metastasis of GC and could be used as a potential target for future treatment of GC.
Keywords: FKBP10, GC, adhesion, lymph node metastasis, integrin
Introduction
Gastric cancer (GC) is one of the most malignant tumor in the world.1 Lymph node metastasis of GC is identified as one of the most important clinicopathological variables which is an important factor affecting the prognosis of patients.2–4 Lymph node metastasis in GC is the result of a combination of multiple driving processes. Current physical examination methods have limited sensitivity and specificity, and there is no effective biomarker for screening lymph node metastasis of GC, which, to a large extent, limits the diagnosis and treatment of GC to a certain extent. Therefore, there is an urgent need to select specific markers that can predict lymph node metastasis in GC to further guide the treatment.
FKBP prolyl isomerase 10 (FKBP10), which can regulate the cross-linking of extracellular matrix collagen, belongs to FK506 binding protein (FKBP65) family.5,6 Thus far, it has been linked with abnormal collagen cross-linking, such as pulmonary fibrosis and tumor.7–11 At present, the role of FKBP10 in tumors is getting more and more attention. Studies have shown that FKBP10 is highly expressed in a variety of tumors, that it promotes tumor progression and is a poor prognostic factor for tumors, such as kidney,12 lung13 and prostate cancer.14 Online data have shown that FKBP10 is a poor prognostic factor for GC, and that it participate in regulating cell adhesion of GC;15 however, its specific mechanisms in GC are still unclear. In this study, we performed differential expression gene screening on four independent GC databases, and verified the prognostic effects of 89 consistently up-regulated genes, finally obtaining 12 prognostic genes. We noted that among these 12 genes, the expression of FKBP10 was significantly elevated in patients with lymph node metastasis. This indicated that FKBP10 might be a specific biomarker for lymph node metastasis of GC, which is associated with poor prognosis of GC. Subsequently basic experiment proved the silencing FKBP10 in GC cells inhibits GC cell adhesion, which is achieved by down-regulating the integrin family of integrin αV and α6.
Materials and Methods
Cell Lines
Human gastric cell lines MKN-45, HGC-27, AGS and the normal human gastric epithelial cell line GES-1 were purchased from the Chinese Academy of Sciences (Shanghai, China). MKN-7 was obtained from the Japanese Collection of Research Bioresources (JCRB Cell Bank, Osaka, Japan). SNU-216 was obtained from the Korea Cell Line Bank (KCLB, Seoul, Korea).
The human gastric cancer cell lines HGC-27 and MKN-7 were used for basic experimental verification. The cell lines were cultured in RPMI 1640 medium containing 10% fetal bovine serum (FBS, Thermo Scientific, MA, USA) and penicillin-streptomycin (100 U/mL-1), and the cells were in a humid environment. It absorbs 95% of air and 5% of CO2 at 37 °C. All experiments were performed in the logarithmic growth phase.
Antibodies and Reagents
Anti-FKBP10 (PA5-63387, 0.4 µg/mL) was purchased from Invitrogen (Rockford, IL, USA). Anti-AKT (9272,1:1000), anti-p-AKT308(4056,1:1000), anti-p-AKT473(9271L,1:1000), anti-integrin αV (4711,1:500) and α5 (4705,1:1000), anti-GAPDH (2118,1:1000) were purchased from Cell Signaling Technology (Danvers, MA, USA). Anti-integrin α6 (ab97760,1:500) was purchased from Abcam (Cambridge, MA, USA). Anti-integrin α1 (mab5676, 2µg/mL) and α2 (mab12331,1µg/mL) were purchased from the R&D system (Minneapolis, MN, USA).
Secondary goat anti-rabbit and goat anti-mouse antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Matrigel was purchased from Corning (Corning Life Science, Tewksbury, MA, USA).
Adhesion Assay
Adhesion assay was performed as previously described.16 HGC-27 and MKN-7 cells transfected with FKBP10 siRNA or NC siRNA were seeded in 96-well plates, which were pre-coated with 10 µg/mL Matrigel at 37 ° C overnight at a density of 1 × 104 cells/200 µL. After incubating at 37 ° C for 30 minutes, wash three times with phosphate buffer saline (PBS) to remove non-adherent cells. Adherent cells were fixed in 4% paraformaldehyde, stained with Wright-Giemsa, then counted at five random fields under a microscope (Olympus) and analyzed statistically.
Western Blot Analysis
After treating cells according to experimental needs. Western blotting was performed as previously described.16 Cells were extracted in 1% Triton lysis buffer and quantified using the Coomassie brilliant blue method. Then, the cell lysates were separated by SDS-PAGE and electrophoretically transferred to a polyvinylidene fluoride (PVDF) membrane (Millipore, Bedford, MA, USA). The membranes were blocked with 5% skim milk in tris-buffered saline Tween-20 (TBST) buffer at room temperature for 1 hour and incubated with the indicated primary antibodies overnight at 4°C, which were diluted proportionally with 5% skim milk in tris-buffered saline Tween-20 (TBST) buffer. After incubating with the appropriate secondary antibodies at room temperature for 40 minutes, the protein bands were detected with enhanced chemiluminescence reagent (SuperSignal Western Pico Chemiluminescent Substrate; Pierce, USA) and visualized with the Electrophoresis Gel Imaging Analysis System (DNR Bio-Imaging Systems, Israel). Final images were analyzed using ImageJ software.
Small Interfering RNA Transfections
After the cell confluence reaches 30% −40%, Lipofectamine 2000 was used to transfect with siRNA according to (Invitrogen, Carlsbad, CA, USA) manufacturer’s instructions. FKBP10 siRNAs and negative control siRNA (NC) were purchased from ViewSolid Biotech (Beijing, China). After 48 hours transfection, efficiency was identified by using RT-PCR and Western blot analysis.
Total RNA Extraction and Real-Time PCR
RT-PCR was performed as previously described.16 Primer sequences for FKBP10: Forward (5ʹ-TGCCTCCCTCAGACTCTAACAC-3ʹ), Reverse (5ʹ-GAATGGGGGCTGGGAAGACTG-3ʹ).
Identification of Differential Expression Genes (DEGs)
We selected four representative GC datasets TCGA-STAD, GSE29272, GSE54129, and GSE27342 for identification of DEGs. The RNAseq matrix and corresponding clinical information of TCGA-STAD cohort were downloaded from TCGA data portal (http://cancergenome.nih.gov/). GSE29272, GSE54129 and GSE27342 were downloaded from the Gene Expression Omnibus (GEO) database. RNA‐seq of TCGA-STAD were normalized using R package “DESeq2”.17 Microarray data of GSE29272, GSE54129 and GSE27342 were normalized and filtered using R package “preprocessCore”. DEGs of TCGA-STAD were screened under the criteria of adj.P < 0.05 and FC ≥ 1.5 using R package “DESeq2”. DEGs of GSE29272, GSE54129, GSE27342 cohort were screened under the criteria of corrected adj.P < 0.05 and FC ≥ 1.5 using R package “limma”.18
Functional Enrichment Analysis of DEGs
In order to understand how stable up-regulated DEGs affect the prognosis of GC patients, we performed KEGG analysis for these DEGs.19 The R package “clusterProfiler” was used to perform gene function enrichment analysis. The difference was considered statistically significant at P < 0.05.
Univariate Cox Regression Analysis
To determine the prognostic value of 89 genes that were consistently up-regulated, we performed a univariate analysis. Univariate analysis was done with the R package "survival" and "survminer". P<0.05 was considered to be meaningful.
Gene Set Enrichment Analysis (GSEA) for FKBP10
We use GSEA v2.2.2 (http://www.broadinstitute.org/gsea) to investigate the biological characteristics of FKBP10.20 Expression level of FKBP10 was annotated as high or low phenotype. C2: CP KEGG gene sets from MSigDB was used as the reference gene sets, all other parameters were set to default.19
Statistical Analysis
The experimental results were analyzed by SPSS 16.0 software. Student’s t-test was used to compare between the two groups. All averages were calculated from three independent experiments. P < 0.05 was considered statistically significant.
Results
Identification of Vital Up-Regulated DEGs in GC
In order to obtain stable DEGs in GC, we performed differential gene analysis on four independent data sets. A total of 3423 up-regulated genes and 3714 down-regulated genes were screened in TCGA-STAD with the criteria: adj.P < 0.05 and FC ≥ 1.5 using R package “DESeq2” (Figure 1G and H). R package “limma” was used for screening DEGs for GSE29272, GSE54129 and GSE27342. Under the criteria that corrected adj.P < 0.05 and FC ≥ 1.5,677 up-regulated genes and 771 down-regulated genes were screened from GSE27342 (Figure 1A and D); 491 up-regulated genes and 359 down-regulated genes were screened from GSE29272 (Figure 1B and E); 1829 up-regulated genes and 1859 down-regulated genes were screened from GSE54129 (Figure 1C and F). A total of 89 genes were simultaneously up-regulated in these 4 datasets (Figure 1I).
KEGG Analysis of the 89-Up Regulated Genes
In order to understand the function of these 89 consistently up-regulated DEGs, KEGG were performed and bubble maps were drawn using ggplot2. KEGG analysis showed that these up-regulated genes are particularly abundant in “protein digestion and absorption,” “PI3K-AKT signal pathway,” and “focal adhesion”signal pathway (Figure 2A). The genes specifically enriched in each KEGG terms are shown in Figure 2B and C. These analysis results indicate that these 89 genes are involved in the classic cancer-promoting pathway of GC, but the specific role needs further clarification.
Identification of FKBP10 as a Novel Prognostic Marker for GC
In order to find genes that participate in the tumorgenesis and progression of GC, we performed single factor analysis of three independent gastric data sets: TCGA, GSE15459, and GSE62254 (Figure 3A and B and C). The obtained results showed that a total of 12 genes exhibited a high expression with poor prognosis in all three datasets (Figure 3D). In this study we mainly explore the factors affecting lymph node metastasis in GC. Further analysis found that only FKBP10 was highly expressed in patients with lymph node metastasis (Figure 3E). Kaplan-Meier analysis found that FKBP10 has stable prognostic effects in three independent gastric cohort (Figure 3F–H). This indicates that FKBP10 can be used not only as a biomarker for the prognosis of GC, but also as a specific biomarker for lymph node metastasis of GC.
Identification of Key Pathways and Genes Related to FKBP10
To further explore how FKBP10 affects GC, we performed GSEA analysis. In three independent GC datasets GSE15459, GSE62254 and TCGA-STAD, “KEGG_ADHERENS_JUNCTION”, “KEGG_ECM_RECEPTOR_INTERACTION” and “KEGG_FOCAL_ADHESION” were all identified as significance pathways related to FKBP10 (Table 1, Figure 4A–C), which makes us presume that FKBP10 may play an important role in the biological behavior of cell adhesion. According to Spearman correlation, we identified 1000 genes that were most relevant to FKBP10. In summary, we speculate that FKBP10 affects the adhesion process of GC through the PI3K-AKT signaling pathway.
 |
Table 1 Top Six GSEA Terms are Significantly Enriched in TCGA-STAD, GSE15459 and GSE62254 |
FKBP10 Promotes GC Cells Adhesion Through Integrin Signaling in vitro
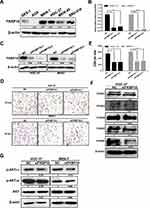
GSEA and KEGG analysis showed that FKBP10 might affect the biological behavior of GC cells by acting on “adhesion pathway” and “PI3K-AKT pathway”. Based on this result, we next use basic experiments to test whether FKBP10 has an effect on the adhesion of GC cells. We first detected FKBP10 protein expression levels in various gastric cancer cell lines and GES-1. Figure 5A shows that FKBP10 is significantly overexpressed in MKN-7 and HGC-27 cell lines, which is significantly higher than GES-1. Then siRNA was used to transiently knock down FKBP10 expression, after transfected with siNC or siFKBP10 RT-PCR and Western Blot were used to detect knockdown efficiency. As shown in Figure 5B and C, siRNA reduced FKBP10 mRNA and protein expression in both cell lines. Adhesion assay results showed FKBP10 knockdown in HGC-27 and MKN-7 cells significantly suppressed cell adhesion compared to the negative controls as shown in Figure 5D and E. This result is consistent with the results of our data analysis. In addition to “adhesion pathway”, data analysis showed that FKBP10 is involved in the“PI3K-AKT pathway” and therefore, it was tested whether FKBP10 affects GC cell adhesion through this pathway. Figure 5G shows that there is no change in the AKT protein after knockdown of FKBP10 in GC cells, while the phosphorylated AKT is reduced. At the same time, we detected changes in expression of integrin family proteins, which are adhesion classical related protein. As shown in Figure 5F, integrin αV and integrin α6 are significantly reduced. These results indicate that FKBP10 has an important role in GC cell adhesion, and that it may act through the integrin/AKT pathway.
Discussion
GC has a high degree of malignancy and a poor prognosis. Many factors including lymph node metastasis can affect the prognosis of GC. Various examination methods have limitations in the diagnosis of gastric lymph node metastasis.21–24 Currently, there are no effective molecular predictors that could be used to predict lymph node metastasis in GC. In this study, we screened out FKBP10 as a potential marker for lymph node metastasis in GC using online data analysis. Our experiments confirmed that GC cell adhesion was down-regulated after silencing FKBP10. Unfortunately, in this study we lacked a classic animal model of orthotopic lymph node metastasis for GC. We will make up for this in future research.
Current research shows that dysfunction of FKBP10 leads to abnormal expression of collagen in the interstitial cells and structural abnormalities, which in turn lead to disease. FKBP10 is up-regulated in multiple solid tumors including lung, liver, and kidney fibrosis cancer.8,9,11 Existing studies have shown that FKBP65 cross-links with pyridine by mediating LH2 dimerization,8 and decreased expression of FKBP10 results in decreased Col1a1 mRNA levels and collagen accumulation in the liver.11 At the same time, FKBP65, HSP47 and Bip form a complex to regulate the activity of LH2.9 Over recent years, the role of FKBP10 in tumors has received increasing attention; nevertheless, its function is controversial. On the one hand, FKBP10 expression is increased in some tumors, and normal tissues are not expressed, such as in melanoma but not in normal tissues.11,25 FKBP10 is not expressed in healthy parenchyma in human but is expressed in lung cancer nevertheless the KRAS is negative or positive. In KRAS-driven lung tumors, down-regulation of FKBP10 inhibits tumor cell growth.13 In colorectal cancer, FKBP10 is highly expressed compared to the corresponding normal tissue.26 Decreased expression of FKBP10 inhibits proliferation and migration of renal cancer cells by down-regulating heat shock protein 90 and resulting in cell cycle arrest.12 In a mouse xenograft model, YK-4-279, which is a small molecule inhibitor of ETV1, could prevent prostate cancer growth and metastasis. This effect is achieved by targeting FKBP10.14 Moreover, in the acquisition and maintenance of the doxorubicin resistance phenotype in leukemia cells FKBP10 also plays an important role.27 However, compared with benign tumor cells and ovarian epithelial cells, the expression of FK506-binding protein 65 (FKBP65) is decreased in epithelial ovarian cancer cells.28 FKBP10 is highly expressed in GC, and is a poor prognostic factor for GC;15 however, the specific mechanism underlying these actions still remain unclear.
In this study, we focus on factors affecting lymph node metastasis (LNM) in GC. We first identified 89 genes that were consistently up-regulated in tumor tissues compared to normal tissues across three GEO database and TCGA-STAD cohort. Out of these 89 genes, only 12 showed a stable prognostic value. FKBP10 was only significantly higher in patients with LNM compared to patients with non-LNM, which implied FKBP10 may promote GC progression by promoting GC lymph node metastasis. Our results revealed that FKBP10 is a potential marker for lymph node metastasis of GC that is associated with poor prognosis of GC. Subsequent GSEA results suggested that FKBP10 might be associated with cell adhesion. In order to verify this, we conducted basic experiments, which confirmed that silencing FKBP10 in GC cells could inhibit GC cell adhesion as was already reported by previous studies. We furthermore found that integrin αV and integrin α6 can be down-regulated after silencing FKBP10. The binding of integrin αV and α6 with their specific ECM portein were downregulation, thus inhibiting their potential downstream P-AKT and GC cell adhesion. This mechanism may promote interstitial enhancement of GC cells, thereby promoting lymph node metastasis.
Conclusion
Compared to non-LNM patients, FKBP10 expression was significantly higher in LNM patients. FKBP10 is a potential marker of lymph node metastasis in GC and is associated with poor prognosis of GC. Online data and basic experiments have verified this, and after FKBP10 is silenced, integrin αV and integrin α6 can be down-regulated, and its classical downstream P-AKT is down-regulated. It is suggested that FKBP10 may promote interstitial enhancement of GC cells through this mechanism, thereby promoting lymph node metastasis.
Acknowledgments
National Science and Technology Major Project of the Ministry of Science and Technology of China (No. 2017ZX09304025); National Natural Science Foundation of China (No. 81972751 and No. 81572374); Science and Technology Plan Project of Liaoning Province (No. 2016007010).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Venerito M, Vasapolli R, Rokkas T, Delchier JC, Malfertheiner P. Helicobacter pylori, gastric cancer and other gastrointestinal malignancies. Helicobacter. 2017;22(Suppl):1. doi:10.1111/hel.12413
2. Hiratsuka M, Iwanaga T, Furukawa H, et al. [Important prognostic factors in surgically treated gastric cancer patients]. Gan to kagaku ryoho. Cancer Chemother. 1995;22(5):703–708.
3. Takagane A, Terashima M, Abe K, et al. Evaluation of the ratio of lymph node metastasis as a prognostic factor in patients with gastric cancer. Gastric Cancer. 1999;2(2):122–128. doi:10.1007/s101200050034
4. Sano T, Coit DG, Kim HH, et al. Proposal of a new stage grouping of gastric cancer for TNM classification: International Gastric Cancer Association staging project. Gastric Cancer. 2017;20(2):217–225. doi:10.1007/s10120-016-0601-9
5. Kang CB, Hong Y, Dhe-Paganon S, Yoon HS. FKBP family proteins: immunophilins with versatile biological functions. Neuro-Signals. 2008;16(4):318–325. doi:10.1159/000123041
6. Patterson CE, Schaub T, Coleman EJ, Davis EC. Developmental regulation of FKBP65. An ER-localized extracellular matrix binding-protein. Mol Biol Cell. 2000;11(11):3925–3935. doi:10.1091/mbc.11.11.3925
7. Ishikawa Y, Vranka J, Wirz J, Nagata K, Bachinger HP. The rough endoplasmic reticulum-resident FK506-binding protein FKBP65 is a molecular chaperone that interacts with collagens. J Biol Chem. 2008;283(46):31584–31590. doi:10.1074/jbc.M802535200
8. Gjaltema RA, van der Stoel MM, Boersema M, Bank RA. Disentangling mechanisms involved in collagen pyridinoline cross-linking: the immunophilin FKBP65 is critical for dimerization of lysyl hydroxylase 2. Proc Natl Acad Sci U S A. 2016;113(26):7142–7147. doi:10.1073/pnas.1600074113
9. Duran I, Martin JH, Weis MA, et al. A chaperone complex formed by HSP47, FKBP65, and BiP modulates telopeptide lysyl hydroxylation of type I procollagen. J Bone Miner Res. 2017;32(6):1309–1319. doi:10.1002/jbmr.3095
10. Chen Y, Terajima M, Banerjee P, et al. FKBP65-dependent peptidyl-prolyl isomerase activity potentiates the lysyl hydroxylase 2-driven collagen cross-link switch. Sci Rep. 2017;7:46021. doi:10.1038/srep46021
11. Vollmann EH, Cao L, Amatucci A, et al. Identification of novel fibrosis modifiers by in vivo siRNA silencing. Mol Ther Nucleic Acids. 2017;7:314–323. doi:10.1016/j.omtn.2017.04.014
12. Ge Y, Xu A, Zhang M, et al. FK506 binding protein 10 is overexpressed and promotes renal cell carcinoma. Urol Int. 2017;98(2):169–176. doi:10.1159/000448338
13. Ramadori G, Konstantinidou G, Venkateswaran N, et al. Diet-induced unresolved ER stress hinders KRAS-driven lung tumorigenesis. Cell Metab. 2015;21(1):117–125. doi:10.1016/j.cmet.2014.11.020
14. Rahim S, Minas T, Hong SH, et al. A small molecule inhibitor of ETV1, YK-4-279, prevents prostate cancer growth and metastasis in a mouse xenograft model. PLoS One. 2014;9(12):e114260. doi:10.1371/journal.pone.0114260
15. Liang L, Zhao K, Zhu JH, Chen G, Qin XG, Chen JQ. Comprehensive evaluation of FKBP10 expression and its prognostic potential in gastric cancer. Oncol Rep. 2019;42(2):615–628. doi:10.3892/or.2019.7195
16. Yan H, Zheng C, Li Z, et al. NPTX1 promotes metastasis via integrin/FAK signaling in gastric cancer. Cancer Manag Res. 2019;11:3237–3251. doi:10.2147/CMAR.S196509
17. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi:10.1186/s13059-014-0550-8
18. Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3(1):Article3. doi:10.2202/1544-6115.1027
19. Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–1740. doi:10.1093/bioinformatics/btr260
20. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi:10.1073/pnas.0506580102
21. Barros RH, Penachim TJ, Martins DL, Andreollo NA, Caserta NM. Multidetector computed tomography in the preoperative staging of gastric adenocarcinoma. Radiol Bras. 2015;48(2):74–80. doi:10.1590/0100-3984.2014.0021
22. Cardoso R, Coburn N, Seevaratnam R, et al. A systematic review and meta-analysis of the utility of EUS for preoperative staging for gastric cancer. Gastric Cancer. 2012;15(Suppl 1):S19–S26. doi:10.1007/s10120-011-0115-4
23. Wu CX, Zhu ZH. Diagnosis and evaluation of gastric cancer by positron emission tomography. World J Gastroenterol. 2014;20(16):4574–4585. doi:10.3748/wjg.v20.i16.4574
24. Cimavilla Roman M, de la Serna Higuera C, Loza Vargas LA, et al. Endoscopic ultrasound versus multidetector computed tomography in preoperative gastric cancer staging. Rev Esp Enferm Dig. 2017;109(11):761–767. doi:10.17235/reed.2017.4638/2016
25. Hagedorn M, Siegfried G, Hooks KB, Khatib AM. Integration of zebrafish fin regeneration genes with expression data of human tumors in silico uncovers potential novel melanoma markers. Oncotarget. 2016;7(44):71567–71579. doi:10.18632/oncotarget.12257
26. Koren J
27. Sun Z, Dong J, Zhang S, et al. Identification of chemoresistance-related cell-surface glycoproteins in leukemia cells and functional validation of candidate glycoproteins. J Proteome Res. 2014;13(3):1593–1601. doi:10.1021/pr4010822
28. Quinn MC, Wojnarowicz PM, Pickett A, et al. FKBP10/FKBP65 expression in high-grade ovarian serous carcinoma and its association with patient outcome. Int J Oncol. 2013;42(3):912–920. doi:10.3892/ijo.2013.1797
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.