Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 14
Ferroptosis-Related Gene Signature: A New Method for Personalized Risk Assessment in Patients with Diffuse Large B-Cell Lymphoma
Authors Chen H, He Y, Pan T, Zeng R, Li Y, Chen S, Li Y, Xiao L, Zhou H
Received 9 March 2021
Accepted for publication 7 May 2021
Published 26 May 2021 Volume 2021:14 Pages 609—619
DOI https://doi.org/10.2147/PGPM.S309846
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Martin H Bluth
Huan Chen,1,* Yizi He,1,* Tao Pan,1 Ruolan Zeng,1 Yajun Li,1 Siwei Chen,2 Yufeng Li,2 Ling Xiao,2 Hui Zhou1
1Department of Lymphoma & Hematology, Hunan Cancer Hospital, The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Changsha, Hunan, People’s Republic of China; 2Department of Histology and Embryology of School of Basic Medical Science, Central South University, Changsha, Hunan, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Hui Zhou; Ling Xiao Email [email protected]; [email protected]
Purpose: Diffuse large B-cell lymphoma (DLBCL) is a genetically heterogeneous disease, which makes prognostic prediction challenging. The rapid development of research on ferroptosis provides the possibility of its use in prognosis in cancer patients. The aim of the current investigation was to perform a systematic study of ferroptosis and DLBCL prognosis to identify prognostic biomarkers in DLBCL.
Materials and Methods: A total of 884 DLBCL patients from the Gene Expression Omnibus database were included in this study and were divided into a training set and a validation set. Univariate Cox regression analysis was used to investigate relationships between gene expression and prognostic values. Ferroptosis-related genes associated with overall survival in the training set were then extracted, and the least absolute shrinkage and selection operator Cox regression model was used to establish an eight-gene signature, comprising ZEB1, PSAT1, NGB, NFE2L2, LAMP2, HIF1A, FH, and CXCL2.
Results: The signature exhibited significant independent prognostic value in both the training set and the validation set. It also exhibited strong prognostic value in subgroup analysis. A nomogram integrating the eight-gene signature and components of the International Prognostic Index facilitated reliable prognostic prediction.
Conclusion: A novel and reliable ferroptosis-related gene signature that can effectively classify DLBCL patients into high-risk and low-risk groups in terms of survival rate was developed. It could be used for prognostic prediction in DLBCL patients. Targeting ferroptosis may be a therapeutic alternative in DLBCL.
Keywords: diffuse large B-cell lymphoma, ferroptosis, Gene Expression Omnibus database, signature, prognostic
Introduction
Diffuse large B-cell lymphoma (DLBCL) is a genetically heterogeneous disease that is divided into germinal center B cell and activated B cell subtypes in transcription.1 The first-line treatment for DLBCL is chemoimmunotherapy using rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP).2 The 5-year survival rate of patients who undergo standard care can reach 60%–70%.3 Approximately 30–40% of patients respond poorly to the standard treatment and mortality is highest within the first 2 years after diagnosis.4 Despite a substantial increase in the investigation of molecular genetics and novel targeted agents in recent years, attempts to improve outcomes by combining standard therapy with novel targeted agents have thus far yielded disappointing results.5 One of the major barriers to the effective use of novel therapies targeting specific pathways is the biological heterogeneity of DLBCL.
In the era of rituximab, the most common international prognostic indicators used in DLBCL patients include the International Prognostic Index (IPI), the Revised IPI, and the National Comprehensive Cancer Network (NCCN-IPI).6 All of these systems use standard clinical and laboratory factors to predict outcomes without explicitly considering the molecular heterogeneity of DLBCL.5,6 Hence, it is becoming increasingly important to define high-risk patients in the early stages of disease and to enter them into clinical trials investigating alternative therapies.
Rapidly developing ferroptosis research suggests the possibility for its use in the formulation of prognoses in cancer patients. Ferroptosis was first proposed by Dixon et al in carcinogen studies.7 It refers to the regulated form of iron-dependent cell death caused by the accumulation of lipid-based reactive oxygen species.8 Studies indicate that a network of long non-coding RNA and competitive endogenous RNA plays an important role in tumorigenesis and ferroptosis in lung cancer.9 The growth of tumor cells can evidently be inhibited by promoting ferroptosis and ferroptosis is also related to tumor resistance.10 Cancer-associated fibroblasts reportedly inhibit the ferroptosis of gastric cancer cells by targeting ALOX15 and blocking the accumulation of lipid-reactive oxygen species(ROS), and secrete exosomal miR-522 thereby promoting drug resistance.11 In a sensitivity analysis conducted using 177 cancer cell lines, lymphoma cell lines were particularly susceptible to GPX4-regulated ferroptosis.12 Erasing an inducer of ferroptosis induces hypertrophy and slows tumor growth in mouse lymphoma models.13
Some basic research suggests that the induction of ferroptosis is a potential therapeutic approach for lymphoma, but there is a lack of research on relationships between clinical lymphoma data and ferroptosis. In the current study the significance of ferroptosis-related mRNA expression was investigated with respect to prognostic evaluation in DLBCL patients. mRNA expression profiles and corresponding clinical data derived from DLBCL lymphoma patients were downloaded from public databases. An analysis of differentially expressed genes related to ferroptosis in the Gene Expression Omnibus cohort was then conducted, and it was subsequently verified in other databases. Lastly, functional enrichment analysis was conducted to investigate potential mechanisms involved. A prognostic model based on a large number of ferroptosis-related genes was thus developed. We believe that this robust prognostic model will improve risk stratification in DLBCL patients and facilitate more accurate assessment for their clinical management.
Materials and Methods
Data Collection
Clinical data and gene expression profiling of patients with DLBCL were obtained from the National Center for Biotechnology Information Gene Expression Omnibus (NCBI Gene Expression Omnibus). Data series were downloaded in a normalized expression matrix file format and used directly in analyses. A training cohort containing 470 patients from the GSE31312 dataset was generated,14 as was a validation cohort containing 414 patients from the GSE10846 dataset.15
Identification of Ferroptosis-Related Genes Associated with Overall Survival
Univariate Cox regression analysis was used to investigate relationships between gene expression and prognostic values, and the results were strengthened by retaining only genes with p values < 0.05. A total of 259 ferroptosis-related genes were retrieved from FerrDb (http://www.zhounan.org/ferrdb). Lastly, ferroptosis-related genes that were significantly associated with overall survival (OS) in the datasets were extracted.
Ferroptosis-Related Risk Model
The GSE31312 dataset was used to construct a ferroptosis-related model. Using least absolute shrinkage and selection operator (LASSO) Cox regression analysis, a risk formula was generated to predict prognoses. The R package “glmnet” statistics software was used to perform the LASSO regression. An individualized risk score was then calculated for each patient, and the median risk score was used as the cut-off to divide all patients into a high-risk group or a low risk-group. Kaplan-Meier survival analysis and Log rank testing were used to evaluate the difference in OS between the high-risk group and the low-risk group. Time-dependent receiver operating characteristic (ROC) curves were constructed to assess prognostic value,16 and relationships between risk scores and clinical characteristics were assessed. Differences in clinical factors between the high-risk and low-risk groups were evaluated via stratified analysis. The analysis was performed using the Sangerbox tool (http://sangerbox.com/Tool).
Univariate and Multivariate COX Regression
SPSS 26 was used for univariate and multivariate COX regression analyses of clinical information and the risk prediction model.
Nomogram Construction and Validation
Based on the GSE31312 cohort, a nomogram integrating ferroptosis-related gene features and IPI components was established to assess the probability of individualized OS at 1, 3, and 5 years using the R package (https://cran.r-project.org/web/packages/rms/). The discriminant capacity of the nomogram was graphically evaluated via calibration mapping.
Functional Enrichment Analyses and Protein-Protein Interaction Network Generation
Gene ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were performed using the “clusterProfiler” R package, then the results were displayed in R language via the ggplot2 package. Adjusted p values < 0.05 were considered statistically significant. A protein–protein interaction (PPI) network was obtained from the STRING online database (https://string-db.org/) and visualized via Cytoscape (version 3.7.2).
Results
Identification of Ferroptosis-Related Genes Associated with OS
The clinical characteristics of all patients in the GSE31312 and GSE10846 datasets are shown in Table 1. Univariate Cox regression was used to assess associations between ferroptosis-related genes and OS. A total of 104 ferroptosis-related genes that were significantly associated with OS in the training dataset were extracted (Supplementary Table S1). A LASSO Cox method was used to select the most useful predictors among the 104 genes in the GSE31312 dataset, and 8 prognostic ferroptosis-related genes with the highest predictive values based on OS were identified (Figure 1A and B).
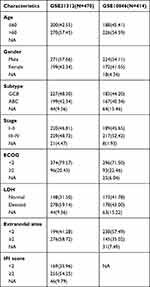 |
Table 1 The Clinical Characteristics of the DLBCL Patients from GEO |
Enrichment Analysis and PPI Networks
To gain insight into the mechanisms ferroptosis-related genes are involved in DLBCL, 104 ferroptosis-related genes were used to perform gene ontology enrichment and KEGG pathway analyses. As expected, in gene ontology enrichment analysis 104 genes were related to oxidative stress-related gene ontology terms, such as “cellular response to oxidative stress” and “response to oxidative stress” (Supplementary Figure S1A). KEGG pathway analysis also indicated that these genes are primarily involved in the ferroptosis pathway (Supplementary Figure S1B). A total of 104 ferroptosis-related genes were used to establish a PPI network (Figure 1C).
Construction of a Prognostic Gene Signature Based on Ferroptosis-Related Gene
LASSO Cox regression was used to determine the optimal weight coefficients for the eight prognostic ferroptosis-related genes, then a prognostic risk score model was established with the following formula:
Risk score = expression level of HIF1A*(−0.05455544) + expression level of NFE2L2*(−1.87952976) + expression level of FH*(−0.09480619) + expression level of LAMP2*(−0.34575386) + expression level of ZEB1*(−0.43828031) + expression level of NGB*(0.16447965) + expression level of PSAT1*(0.47117607) + expression level of CXCL2*(−0.20840707)
Risk scores were calculated for all 470 patients in the training set, who were then assigned to a high-risk group or a low-risk group based on whether their risk score was higher or lower than the median risk score. The distributions of risk scores, death status, survival time, and expression levels of the eight genes used to construct the prognostic signature were visualized to evaluate prognostic differences between the two groups (Figure 2A–C). The expression of 8 genes was different between high and low risk groups (Supplementary Figure S2). The respective areas under time-dependent ROC curves of the eight-gene model at 1, 3, and 5 years were 0.71, 0.66, and 0.66 (Figure 2D), indicating that the model had high sensitivity and high specificity with regard to predicting the prognosis of DLBCL patients. In Kaplan-Meier curve analysis, patients with high-risk scores had significantly poorer OS than patients with low-risk scores (hazard ratio 8.62; 95% confidence interval 5.39–13.77; p < 0.0001) (Figure 2E). In order to explore the correlation among the eight core genes, we constructed a PPI network, and the results showed that six genes out of eight were closely related (Supplementary Figure S1C).
Relationships Between Risk Score and Clinical Features
Correlations between risk score and age, Eastern Cooperative Oncology Group (ECOG), IPI score, lactate dehydrogenase (LDH), stage, and extranodal sites were calculated. Risk score was positively correlated with age (Figure 3A), IPI score (Figure 3C), LDH (Figure 3D), DLBCL stage (Figure 3E), and extranodal sites (Figure 3F), but not with ECOG (Figure 3B).
 |
Figure 3 Correlations between risk score and age (A), ECOG (B), IPI score (C), LDH (D), DLBCL stage (E), extranodal sites (F).*p < 0.05, ***p < 0.001, ****p < 0.0001, ns, p > 0.05. |
Subgroup Analysis of the Eight-Gene Signature in the Training Dataset
The prognostic capacity of the eight-gene signature was further validated in DLBCL subgroups. The signature could predict survival outcomes in clinical subgroups defined by parameters such as activated B cell or germinal center B cell subtype, clinical stage, and IPI score. Patients in the high‐risk group exhibited poorer prognoses irrespective of activated B cell or germinal center B cell subtype (Figure 4A and D), clinical stage (Figure 4B and E), and IPI score (Figure 4C and F). Thus, the signature exhibited independent predictive capacity in clinical application.
Validation of the Eight-Gene Signature in Patients in the GSE10846 Dataset
In the GSE10846 dataset patients classified as low-risk based on the median risk score had longer OS than patients classified as high-risk (hazard ratio 2.1; 95% confidence interval 1.50–2.93; p < 0.0001) (Figure 5A). Respective areas under the ROC curve for predicting OS at 1, 3, and 5 years were 0.610, 0.654, and 0.647, respectively (Figure 5B).
Univariate and Multivariate COX Regression
In analyses performed solely on datasets with IPI components (GSE31312 and GSE10846), the eight-gene signature was an independent prognostic indicator (Table 2).
 |
Table 2 Univariable and Multivariable Cox Regression Analysis in DLBCL |
Construction and Validation of the Nomogram
IPI components and the eight-gene model were used to create nomograms to predict 1, 3, and 5-year survival (Figure 6A). A calibration chart indicated acceptable agreement between the predicted survival rate and the actual survival rate, suggesting that the nomogram may reliably predict DLBCL patient prognoses in clinical practice (Figure 6B–D).
Discussion
DLBCL is a distinct disease, but exhibits extensive molecular heterogeneity and different clinical manifestations, morphological characteristics, immunophenotypes, molecular subsets, and clinical prognoses.17 DLBCL patient risk stratification and prognosis remain a challenge for clinicians. In the rituximab era it was found that IPI was somewhat deficient with respect to distinguishing risk groups, especially high-risk groups, and its prognostic value was found to be lacking.6 The study formulated the Revised-IPI, which divides patients treated via the R-CHOP regimen into three groups. This overcame the problem that the survival curves of patients in low-risk, low-medium-risk, medium-high-risk, and high-risk groups merged.6 Notably however, there was still an inability to adequately identify high-risk patients. Studies indicate that NCCN-IPI classification can quantify the degree of patient risk to an extent. However, all three scoring systems fail to identify high risk group with long-term OS clearly below 50%.6 Moreover, the progression of tumors is affected by numerous biological parameters in addition to those ascertained via clinical and laboratory investigations.
Ferroptosis may have a profound impact on carcinogenesis and prognosis. It is an iron-dependent non-apoptotic form of cell death that can inhibit tumor growth and increase the sensitivity of chemotherapy.18 Multiple small molecular compounds have been identified that induce ferroptosis by targeting iron metabolism and lipid peroxidation,19 but the role and significance of mRNA expression of ferroptosis-related genes in the prognostic assessment of DLBCL are currently unclear. In the current study a prognostic signature based on eight markers that strongly and reliably predicted individualized prognoses in DLBCL patients on the basis of ferroptosis-related genes was established. In conjunction with the incorporation of an IPI component a comprehensive prognosis nomogram was developed that exhibited a good capacity to accurately predict prognoses and positive benefit of DLBCL.
The eight gene products included in the signature developed in the current study were ZEB1, PSAT1, NGB, NFE2L2, LAMP2, HIF1A, FH, and CXCL2. ZEB1 is the main component of the transcription factor network controlling epithelial-to-mesenchymal transition, which is associated with tumor growth and metastasis of malignant tumors via effects on critical cellular functions and states including differentiation, proliferation, responses to DNA damage, and cell survival.20 c-Myc is a common transcription factor of epithelial–mesenchymal transition (EMT) including Snail, ZEB1, and ZEB2 via direct associations with their promoter regions containing E-box motifs, suggesting close correspondence between c-Myc and ZEB1 in the regulation of EMT.21 In DLBCL, ZEB1 activates SNHG14 and PD-L1, which in turn promotes immunoevasion by certain tumors.22 Recent research suggests that ZEB1 overexpression increases susceptibility to ferroptosis, whereas ZEB1 silencing decreases susceptibility to it.23 Many studies indicate that NRF2 plays an important role in the therapeutic response to ferroptosis-targeted therapies in hepatocellular carcinoma.24,25 In signal pathway, NRF2/CBS signal transduction confers elastin-induced ferroptosis resistance.26 The NRF2-Keap1 pathway is essential for cancer cell growth.27 Interaction between ARF and NRF2 is critical for p53-independent ferroptosis, and inhibition of the NRF2-ARE pathway reversed resistance to ferroptosis in head and neck cancer.28 NRF2 is reportedly an effective therapeutic target for chemotherapeutic drugs to make cancer sensitive.25 The hypoxia-inducible factor (HIF) pathway was the driving factor for the vulnerability of ferroptosis.29 That study demonstrated that HIF1A can act as a tumor promoter by degrading p53 protein via binding to five response elements in the p53 promoter, and increases their invasive and metastatic activity.30 HIF1A also activates autophagy, a lysosomal degradation pathway that may promote DLBCL cell survival.31 PSAT1 is upregulated in Burkitt lymphoma via a MYC-dependent ATF4 transcription factor mechanism, and overexpression of PSAT1 is reportedly associated with poor prognoses in patients with Burkitt lymphoma,32 nasopharyngeal carcinoma,33 colorectal carcinoma,34,35 and epithelial ovarian cancer.36 Studies suggest that NGB may be a tumor suppressor that inhibits cell growth, cell aggregation, and tumorigenicity.37 LAMP2 deficiency increases the risk of ferroptosis in retinal pigment epithelial cells.38 ZEB1 and HIF1A can reportedly influence ferroptosis in DLBCL, but the molecular functions of other ferroptosis-related genes in DLBCL patients remain to be elucidated.
The current study had some limitations. Due to the unavailability of control group samples in the Gene Expression Omnibus databases and the retrospective study design, there was a degree of bias. Thus, a well-designed, international, prospective, multicenter clinical trial is needed to validate our findings in the future. In addition, further functional studies are warranted to explore the molecular functions of the identified ferroptosis-related genes during DLBCL progression.
Conclusion
This is the first time a ferroptosis-related prognostic model has been established in DLBCL patients that is capable of serving as a promising prognostic indicator in combination with clinical IPI components. The model may also facilitate the development of new clinical ferroptosis-targeted therapies in patients with DLBCL.
Data Sharing Statement
Publicly available datasets were analyzed in this study. These data can be found at https://www.ncbi.nlm.nih.gov/geo/.
Ethics Statement
This article does not contain any studies with human participants performed by any of the authors.
Consent for Publication
All authors have read the final manuscript and approve of the publication of its content in entirety.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (grant number 82000200); the Natural Science Foundation of Hunan Provincial Health Commission (grant number 20201659); the National Cancer Center (grant number NCC2017A20); the “Scientific Research Climbing Plan” of Hunan Cancer Hospital (grant number ZX2020003); the Whiteknit Foundation’s Vairocoda Hematology Development Program (grant number WAKD008), and Fundamental Research Funds for the Central Universities of Central South University (grant numbers 2019zzts1002 and 2019zzts1060).
Author Contributions
Author Contributions Huan Chen, Yizi He wrote the manuscript; Hui Zhou, Ling Xiao designed the research; Tao Pan, Ruolan Zeng, Yajun Li performed the research; Siwei Chen and Yufeng Li writing-review and editing. All authors have participated in the design, data acquisition, or data analysis and interpretation of this study and made significant contributions. All participated in drafting articles or making strict revisions to important knowledge content; all agreed to submit to the current journal; finally approved the version to be published; and agreed to be responsible for all aspects of the work.
Disclosure
The authors report that there are no conflicts of interest associated with this work.
References
1. Rosenwald A, Wright G, Chan WC, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346(25):1937–1947. doi:10.1056/NEJMoa012914
2. Pfreundschuh M, Kuhnt E, Trümper L, et al. CHOP-like chemotherapy with or without rituximab in young patients with good-prognosis diffuse large-B-cell lymphoma: 6-year results of an open-label randomised study of the MabThera International Trial (MInT) Group. Lancet Oncol. 2011;12(11):1013–1022. doi:10.1016/S1470-2045(11)70235-2
3. Coiffier B, Thieblemont C, Van Den Neste E, et al. Long-term outcome of patients in the LNH-98.5 trial, the first randomized study comparing rituximab-CHOP to standard CHOP chemotherapy in DLBCL patients: a study by the Groupe d’Etudes des Lymphomes de l’Adulte. Blood. 2010;116(12):2040–2045. doi:10.1182/blood-2010-03-276246
4. Yin X, Xu A, Fan F, et al. Incidence and mortality trends and risk prediction nomogram for extranodal diffuse large B-Cell lymphoma: an analysis of the surveillance, epidemiology, and end results database. Front Oncol. 2019;9:1198. doi:10.3389/fonc.2019.01198
5. Link BK. Foreseeing what is to happen in DLBCL. Blood. 2020;135(23):2014–2015. doi:10.1182/blood.2020005678
6. Ruppert AS, Dixon JG, Salles G, et al. International prognostic indices in diffuse large B-cell lymphoma: a comparison of IPI, R-IPI, and NCCN-IPI. Blood. 2020;135(23):2041–2048. doi:10.1182/blood.2019002729
7. Yang WS, SriRamaratnam R, Welsch ME, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156(1–2):317–331. doi:10.1016/j.cell.2013.12.010
8. Dixon SJ, Lemberg KM, Lamprecht MR, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. doi:10.1016/j.cell.2012.03.042
9. Wang M, Mao C, Ouyang L, et al. Long noncoding RNA LINC00336 inhibits ferroptosis in lung cancer by functioning as a competing endogenous RNA. Cell Death Differ. 2019;26(11):2329–2343. doi:10.1038/s41418-019-0304-y
10. Yu H, Guo P, Xie X, Wang Y, Chen G. Ferroptosis, a new form of cell death, and its relationships with tumourous diseases. J Cell Mol Med. 2017;21(4):648–657. doi:10.1111/jcmm.13008
11. Zhang H, Deng T, Liu R, et al. CAF secreted miR-522 suppresses ferroptosis and promotes acquired chemo-resistance in gastric cancer. Mol Cancer. 2020;19(1):43. doi:10.1186/s12943-020-01168-8
12. Kinowaki Y, Kurata M, Ishibashi S, et al. Glutathione peroxidase 4 overexpression inhibits ROS-induced cell death in diffuse large B-cell lymphoma. Lab Invest. 2018;98(5):609–619. doi:10.1038/s41374-017-0008-1
13. Zhang Y, Tan H, Daniels JD, et al. Imidazole ketone erastin induces ferroptosis and slows tumor growth in a mouse lymphoma model. Cell Chem Biol. 2019;26(5):623–633.e629. doi:10.1016/j.chembiol.2019.01.008
14. Visco C, Li Y, Xu-Monette ZY, et al. Comprehensive gene expression profiling and immunohistochemical studies support application of immunophenotypic algorithm for molecular subtype classification in diffuse large B-cell lymphoma: a report from the International DLBCL Rituximab-CHOP Consortium Program Study. Leukemia. 2012;26(9):2103–2113. doi:10.1038/leu.2012.83
15. Lenz G, Wright G, Dave SS, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008;359(22):2313–2323. doi:10.1056/NEJMoa0802885
16. Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56(2):337–344. doi:10.1111/j.0006-341X.2000.00337.x
17. Lacy SE, Barrans SL, Beer PA, et al. Targeted sequencing in DLBCL, molecular subtypes, and outcomes: a haematological malignancy research network report. Blood. 2020;135(20):1759–1771. doi:10.1182/blood.2019003535
18. Liang C, Zhang X, Yang M, Dong X. Recent progress in ferroptosis inducers for cancer therapy. Adv Mater. 2019;31(51):e1904197. doi:10.1002/adma.201904197
19. Stockwell BR, Friedmann Angeli JP, Bayir H, et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171(2):273–285. doi:10.1016/j.cell.2017.09.021
20. Caramel J, Ligier M, Puisieux A. Pleiotropic roles for ZEB1 in cancer. Cancer Res. 2018;78(1):30–35. doi:10.1158/0008-5472.CAN-17-2476
21. Larsen JE, Nathan V, Osborne JK, et al. ZEB1 drives epithelial-to-mesenchymal transition in lung cancer. J Clin Invest. 2016;126(9):3219–3235. doi:10.1172/JCI76725
22. Zhao L, Liu Y, Zhang J, Liu Y, Qi Q. LncRNA SNHG14/miR-5590-3p/ZEB1 positive feedback loop promoted diffuse large B cell lymphoma progression and immune evasion through regulating PD-1/PD-L1 checkpoint. Cell Death Dis. 2019;10(10):731. doi:10.1038/s41419-019-1886-5
23. Lee J, You JH, Kim MS, Roh JL. Epigenetic reprogramming of epithelial-mesenchymal transition promotes ferroptosis of head and neck cancer. Redox Biol. 2020;37:101697. doi:10.1016/j.redox.2020.101697
24. Sun X, Ou Z, Chen R, et al. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology. 2016;63(1):173–184. doi:10.1002/hep.28251
25. Zimta AA, Cenariu D, Irimie A, et al. The role of Nrf2 activity in cancer development and progression. Cancers. 2019;11(11):1755. doi:10.3390/cancers11111755
26. Liu N, Lin X, Huang C. Activation of the reverse transsulfuration pathway through NRF2/CBS confers erastin-induced ferroptosis resistance. Br J Cancer. 2020;122(2):279–292. doi:10.1038/s41416-019-0660-x
27. Fan Z, Wirth AK, Chen D, et al. Nrf2-Keap1 pathway promotes cell proliferation and diminishes ferroptosis. Oncogenesis. 2017;6(8):e371. doi:10.1038/oncsis.2017.65
28. Chen D, Tavana O, Chu B, et al. NRF2 is a major target of ARF in p53-independent tumor suppression. Mol Cell. 2017;68(1):224–232 e224. doi:10.1016/j.molcel.2017.09.009
29. Tiwari A, Tashiro K, Dixit A, et al. Loss of HIF1A from pancreatic cancer cells increases expression of PPP1R1B and degradation of p53 to promote invasion and metastasis. Gastroenterology. 2020;159(5):1882–1897 e1885. doi:10.1053/j.gastro.2020.07.046
30. Madan E, Parker TM, Pelham CJ, et al. HIF-transcribed p53 chaperones HIF-1alpha. Nucleic Acids Res. 2019;47(19):10212–10234. doi:10.1093/nar/gkz766
31. Bhalla S, Evens AM, Prachand S, Schumacker PT, Gordon LI. Paradoxical regulation of hypoxia inducible factor-1α (HIF-1α) by histone deacetylase inhibitor in diffuse large B-cell lymphoma. PLoS One. 2013;8(11):e81333. doi:10.1371/journal.pone.0081333
32. Bialopiotrowicz E, Noyszewska-Kania M, Kachamakova-Trojanowska N, et al. Serine biosynthesis pathway supports MYC-miR-494-EZH2 feed-forward circuit necessary to maintain metabolic and epigenetic reprogramming of burkitt lymphoma cells. Cancers. 2020;12(3):580. doi:10.3390/cancers12030580
33. Liao KM, Chao TB, Tian YF, et al. Overexpression of the PSAT1 gene in nasopharyngeal carcinoma is an indicator of poor prognosis. J Cancer. 2016;7(9):1088–1094. doi:10.7150/jca.15258
34. Fang Y, Liang X, Xu J, Cai X. miR-424 targets AKT3 and PSAT1 and has a tumor-suppressive role in human colorectal cancer. Cancer Manag Res. 2018;10:6537–6547. doi:10.2147/CMAR.S185789
35. Qian C, Xia Y, Ren Y, Yin Y, Deng A. Identification and validation of PSAT1 as a potential prognostic factor for predicting clinical outcomes in patients with colorectal carcinoma. Oncol Lett. 2017;14(6):8014–8020. doi:10.3892/ol.2017.7211
36. Zhang Y, Li J, Dong X, et al. PSAT1 regulated oxidation-reduction balance affects the growth and prognosis of epithelial ovarian cancer. Onco Targets Ther. 2020;13:5443–5453. doi:10.2147/OTT.S250066
37. Lee H, Kim D, Dan HC, et al. Identification and characterization of putative tumor suppressor NGB, a GTP-binding protein that interacts with the neurofibromatosis 2 protein. Mol Cell Biol. 2007;27(6):2103–2119. doi:10.1128/MCB.00572-06
38. Lee JJ, Ishihara K, Notomi S, et al. Lysosome-associated membrane protein-2 deficiency increases the risk of reactive oxygen species-induced ferroptosis in retinal pigment epithelial cells. Biochem Biophys Res Commun. 2020;521(2):414–419. doi:10.1016/j.bbrc.2019.10.138
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.