Back to Journals » Clinical Epidemiology » Volume 10
Feasibility of salivary DNA collection in a population-based case–control study: a pilot study of pediatric Crohn’s disease
Authors Kappelman MD, Lange A , Randell RL, Basta PV, Sandler RS, Laugesen K , Byrjalsen A , Christensen T , Frøslev T, Erichsen R
Received 6 June 2017
Accepted for publication 5 October 2017
Published 28 February 2018 Volume 2018:10 Pages 215—222
DOI https://doi.org/10.2147/CLEP.S143322
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 7
Editor who approved publication: Professor Vera Ehrenstein
Michael D Kappelman,1 Aksel Lange,2 Rachel L Randell,3 Patricia V Basta,4 Robert S Sandler,5 Kristina Laugesen,2 Anna Byrjalsen,2 Tina Christensen,2 Trine Frøslev,2 Rune Erichsen2,6
1Department of Pediatrics, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA; 2Department of Clinical Epidemiology,Aarhus University Hospital, Aarhus, Denmark; 3Department of Pediatrics, Duke University School of Medicine, Duke University, Durham, NC, USA; 4Department of Epidemiology, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA; 5Department of Medicine, University of North Carolina at Chapel Hill, Chapel Hill, NC, USA; 6Surgical Department, Horsens Regional Hospital, Horsens, Denmark
Background: Epidemiologic studies combining exposure and outcome data with the collection of biosamples are needed to study gene–environment interactions that might contribute to the etiology of complex diseases such as pediatric Crohn’s disease (CD). Nationwide registries, including those in Denmark and other Scandinavian countries, provide efficient and reliable sources of data for epidemiological studies evaluating the environmental determinants of disease. We performed a pilot study to test the feasibility of collecting salivary DNA to augment registry data in established cases of pediatric CD and randomly selected, population-based controls.
Subjects and methods: Cases of CD born after 1995 and residing in the central region of Denmark were identified through the Danish National Patient Registry and confirmed by using standard diagnostic criteria. Age- and gender-matched controls were selected at random through the civil registration system. Cases and controls were contacted by mail and telephone and invited to submit a saliva sample. DNA was extracted and genotyped for six CD-associated single-nucleotide polymorphisms (SNPs).
Results: A total of 53 cases of pediatric CD were invited, and 40 contributed a saliva sample (75% response rate). A total of 126 controls were invited, and 54 contributed a saliva sample (44% response rate). As expected, demographic characteristics did not differ between cases and controls. DNA was successfully isolated from 93 of 94 samples. Genotyping was performed with only 2% undetermined genotypes. For five of six SNPs known to be associated with CD, risk allele frequencies were higher in cases than controls.
Conclusion: This pilot study strongly supports the feasibility of augmenting traditional epidemiological data from Danish population-based registries with the de novo collection of genetic information from population-based cases and controls. This will facilitate rigorous studies of gene–environment interactions in complex chronic conditions such as CD.
Keywords: Crohn’s disease, children, genetic, DNA, SNP
Background
Crohn’s disease (CD), believed to affect nearly 10,000 Danes1 and 600,000 Americans,2 is a chronic, idiopathic, inflammatory bowel disease (IBD) that results in substantial morbidity, including frequent hospitalization and surgery,3,4 missed work5 and school,6 and reductions in quality of life.7,8 Nearly 10% of cases are diagnosed during childhood and adolescence, and early-onset IBD is considered to be a particularly severe phenotype. The pathogenesis is believed to involve dysregulation of the gastrointestinal immune response to environmental factors, such as commensal enteric bacteria.9 The genetic predisposition for this condition, long suggested by the approximately 50% concordance in identical twins,10,11 has received further support over the past few years as recent genome-wide association studies have revealed at least 163 susceptibility loci for IBD, emphasizing the range and complexity of pathways that may be involved.12
Despite these strong genetic influences, evidence also suggests a critical role of environmental factors. First, about 50% of monozygotic twins are discordant for CD, despite identical genetic makeup. Second, incidence rates have risen dramatically between 1960 and 1990 in a number of North American and European populations, more than would be expected based on Mendelian inheritance alone.13 In addition, the incidence of IBD has increased exponentially in formerly low-incidence areas including Asia and Latin America, possibly reflecting the “westernization” of these regions including changes in diet, lifestyle, medication use, and perhaps other environmental exposures.13 Nevertheless, studies examining the role of specific environmental risk factors (with the exception of tobacco smoke) have been disappointing, yielding either contradictory or inconclusive results, perhaps due to design limitations resulting in recall and/or referral biases and the lack of study of gene–environment interactions. In complex conditions such as CD, gene–environment interactions have been hypothesized, as the effects of environmental exposures may be dependent on genetic predisposition.14
The ideal study design to evaluate the etiology of a complex, chronic illness that likely involves gene–environment interactions, such as CD, would be a population-based study that combines the measurement of environmental exposures with the collection of genetic factors. Denmark, with a population of some 5 million people, is well suited for epidemiological studies designed to evaluate both the genetic and environmental determinants of complex illnesses such as childhood CD. Since the 1930s, the Danish government has compiled hundreds of databases to monitor the demographics, health, occupational status, and other socioeconomic factors affecting its population. Importantly, each citizen has a unique personal identification number that can be used to tie these individual databases together and track individuals across their lifespan.15,16 Furthermore, the personal identification number can also be used to locate individuals and solicit participation in epidemiological studies requiring additional primary data collection, and/or collection of biological specimens. This infrastructure makes it theoretically possible to conduct methodologically rigorous population-based epidemiological studies that combine the use of existing, prospectively collected administrative data with collection and banking of biological samples.
While the collection and banking of DNA from blood samples of clinical populations (cases) has been common practice for decades, the ability to collect DNA from randomly selected, population-based controls presents a logistical challenge. However, in recent years, the ability to extract DNA from saliva samples has made it theoretically possible for both cases and controls to collect saliva in the comfort of their own home and ship the samples by mail for subsequent extraction and analyzing of DNA. This would eliminate the need for venipuncture and/or study visits, important barriers to recruitment in population-based studies. Whether cases and population-based controls would be willing and able to provide a salivary DNA sample, especially in childhood, remains unknown. We therefore performed a pilot and feasibility study to test methods for the collection, storage, extraction, and genotyping of salivary DNA in confirmed cases of pediatric CD along with age- and gender-matched population controls. If feasible, this would lay the foundation for large, population-based studies of gene–environment interactions across a range of complex chronic conditions in children and adults.
Subjects and methods
Case recruitment
We performed a pilot study to collect and analyze salivary DNA from population-based cases of pediatric CD and age- and gender-matched controls. We identified cases born after 1995 and residing in the central region of Denmark through the Danish National Patient Registry using Crohn’s specific ICD-10 codes K50. The Danish National Patient Registry has kept records of all nonpsychiatric discharge diagnoses since 1977 and outpatient visits since 1995.17 All CD cases identified through this registry were reviewed by a pediatric gastroenterologist (AL) using standard diagnostic criteria.18 Patients without a confirmed diagnosis of CD were excluded.
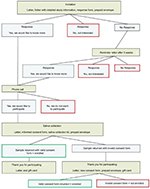
An overview of study recruitment processes is shown in Figure 1. The parents of confirmed cases were mailed a letter describing the study and offered more information about the study participation. If a response form was not returned to the study team within 3 weeks, a reminder letter was sent to the parents. Parents who returned a response form stating they would like to know more about the study were contacted by telephone by a pediatric gastroenterologist (AL) to discuss the purpose and procedures of the study and answer any questions. The parents of cases were also provided additional study-related information during study, office appointments. Parents were then mailed a written consent form along with a salivary DNA collection kit (Oragene; DNA Genotek, Ottawa, ON, Canada), self-collection instructions, and a prepaid return envelope. We utilized Oragene OG-500 kits for saliva collection, as this method has been previously demonstrated to provide high-quality genomic DNA.19 Upon collection, the saliva sample is stable at room temperature for up to 1 year, eliminating the need for immediate laboratory extraction. Participants were instructed not to eat, drink, smoke, or chew gum 30 minutes before sample collection. Upon collection, samples were mailed at ambient temperature without ice to the Department of Clinical Epidemiology at Aarhus University and subsequently batch shipped to the University of North Carolina at Chapel Hill for DNA extraction and analysis. As an incentive for participation, participants were mailed a gift card worth 100 Danish Krone (DKK) (approximately 15 USD) once they had returned a saliva sample.
For each case, we selected age-, gender-, and region-matched controls at random through the civil registration system. To ensure that controls were truly population based, there were no exclusion criteria for controls. As with cases, the parents of selected controls were mailed a letter describing the study and offered additional information about the study participation. A reminder letter was sent as necessary. Parents who returned a response form stating they would like to know more about the study were contacted by telephone by a trained member of the study team to discuss the study in more detail and were then mailed a written consent form along with a salivary DNA collection kit. Controls received the same participation incentive as cases.
DNA extraction and genotyping
All salivary DNA collection kits returned with a valid, signed informed consent form were batch shipped to the University of North Carolina at Chapel Hill for DNA extraction, storage, and analysis.
DNA was extracted from saliva samples using the Chemagic Magnetic Separation Module I (MSMI) robotic system (PerkinElmer Inc., Waltham, MA, USA), using the Chemagic DNA Saliva Kit and the MSMI 24 rod head. The MSMI system isolates DNA after cell lysis via highly specific binding of the DNA to proprietary magnetic particles based on polyvinyl alcohol (M-PVA) magnetic beads. Once bound, the DNA was washed several times and then released from the magnetic beads. Optical density readings were taken on a Nanodrop to assess the 260/280 and 260/230 ratio quality metrics. DNA quantitation was assessed via Picogreen using the Quant-iT PicoGreen dsDNA Assay Kit cat# P7589 (Thermo Fisher Scientific, Waltham, MA, USA).
DNA was genotyped for six CD-associated single-nucleotide polymorphisms (SNPs) using TaqMan SNP Genotyping Assays from Thermo Fisher Scientific. SNPs of interest were selected by first performing a literature search for loci of interest for CD, ulcerative colitis (UC), or IBD,12,20–25 and findings were narrowed to loci that demonstrated relatively large proportion of variance or relatively high odds ratios for IBD versus control.12 Selected SNPs included those for NOD2 (rs2066844, rs2066845, and rs2066847), ATG16L1 (rs12994997), IL23R (rs11209026), and MUC 19 (rs11564258).12,26,27 We used pre-designed assays for all but one SNP (rs2066847), for which a custom primer was designed using previously established sequences (forward primer: GTCCAATAACTGCATCACCTACCT; reverse primer: CAGACTTCCAGGATGGTGTCATTC, probe 1 – VIC-MGB; dye: CAGGCCCCTTGAAAG, probe 2 – FAM-MGB; dye: CAGGCCCTTGAAAG).28 Polymerase chain reaction (PCR) volume was 5 mL.
Sample size
Our targeted sample size was approximately 50 cases and 50 controls to provide a rough estimate of the feasibility of case and control recruitment and to allow accurate genotyping for SNPs with low minor allele frequencies.
Statistical analysis
We used descriptive statistics and Student’s t-tests or Fisher’s exact test as applicable for comparisons between groups. As this was a pilot study, we specifically did not perform statistical comparisons for genetic analyses. All statistics were computed using SAS version 9.3. The study protocol was approved by the Danish Data Protection Agency (J.no. 2010-41-4888), the Scientific Ethical Board of Central Denmark Region (J. no. 1-10-72-372-12), and the University of North Carolina at Chapel Hill.
Results
Study population
A total of 53 cases of pediatric CD were invited, and 40 contributed a saliva sample along with valid consent (75% response rate). The mean age of cases who contributed a sample was 15 years (SD=2.4); 35% were female. The mean age of nonresponder cases was 14.9 years; 54% were female. A total of 126 controls were invited, and 54 contributed a saliva sample along with a valid consent (44% response rate). The mean age of controls who contributed a sample was 15.2 years (SD=2.4); 37% were female. The mean age of nonresponder cases was 15.8 years; 23% were female. As expected, demographics did not differ between cases and controls (Table 1).
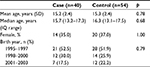  | Table 1 Demographics of pediatric CD cases and controls Abbreviation: CD, Crohn’s disease. |
DNA extraction and yield
Of 94 samples received, one leaked (case sample) in transport. DNA was successfully isolated from the 93 remaining samples. The total DNA yield ranged from 3.11 to 100.34 mg (median 58.20 mg), and 89% (83/93) of the samples yielded >20 mg.
Genotyping
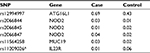
All samples were genotyped for six SNPs associated with IBD, and risk allele frequencies (RAFs) were calculated. Of 564 possible genotypes, only 11 (2.0%) were undetermined. The RAFs for cases and controls are provided in Table 2. Genotypes are provided in Table S1. For five of six SNPs analyzed (83%), the RAFs in cases and controls were all in the expected direction based on prior literature. The only exception was for rs206845 whose RAF was 0.01 in cases and 0.02 in controls.
Discussion
Epidemiologic studies combining exposure and outcome data with the collection of biosamples are needed to study gene–environment interactions that might contribute to the etiology of complex diseases such as pediatric CD. Nationwide registries, including those in Denmark and other Scandinavian countries, provide efficient and reliable sources of data for epidemiological studies evaluating the environmental determinants of disease. In this pilot study, we have demonstrated the capacity to augment registry data through the collection of salivary DNA by mail in both cases, and most importantly, valid, randomly selected, population-based controls. This includes home saliva collection, shipping, storage, DNA extraction, and genotyping.
Specifically, we found a very high response rate of 75% among cases of pediatric CD, and a reasonably high response rate of 44% for randomly selected, age- and gender-matched controls. DNA was successfully extracted from 93 of 94 collected samples, with a median yield of 58.20 mg of DNA per sample. Genotyping was successfully performed on all samples. Overall, only 2% of all genotypes were undetermined. In addition, even in this small pilot study, we were able to replicate previously established genetic associations for five of the six selected genotypes, further demonstrating the utility of augmenting population-based registry studies with genetic analyses.
The response rates observed in this study warrant particular consideration. First, the 75% response rate among CD patients was outstanding. To our knowledge, this is the only study of the response rate for a population-based genetic study of a pediatric illness. However, our response rate was similar to that observed in other highly engaged populations, including participants in a Danish nursing cohort study29 and a US-based cohort of adult patients with IBD.30 We believe that the high response rate among cases likely reflects a patient population that is highly motivated to contribute IBD research for both personal and altruistic reasons.31 In addition, we utilized office visits, in addition to mail/telephone to contact potential participants and provide them with information about the study. Future genetic epidemiology studies may also benefit by augmenting mail/telephone recruitment with office-based contact.
The response rate of 44% among randomly selected controls is also notable. While the expected response rate of randomly selected, population-based, pediatric controls to a genetic study is unknown, our observed response rate seems quite robust given that the response rates to survey/interview studies in adults are in the range of 42%–64%,32–34 and one might have expected a lower response rate for a genetic study in children. Furthermore, we believe the possibility of nonresponse bias is likely to be low in studies of gene–environment interactions, as this would require a gene associated with both nonresponse and disease susceptibility.
The strengths of this study include the use of a population-based registry to identify cases and randomly select potential controls and the confirmation of all cases by an experienced pediatric gastroenterologist. An additional strength was the use of a centralized approach to contact potential controls using only mail and telephone, as this approach can be readily scaled to facilitate large, population-based studies. As with all pilot projects, the main limitation of this work was the limited number of participants which precluded our ability for hypothesis testing of genetic associations between cases and controls. In addition, while our recruitment target was 50 cases, we fell short of this as only 40 cases contributed a sample with valid consent. Another possible limitation is that some patients with CD (diagnosed or undiagnosed) did not have the relevant ICD-10 diagnosis code recorded in the Danish National Registry of Patients (DNRP) and may have been misclassified as controls. However, given the low prevalence of pediatric CD, the potential for misclassification of cases as controls is extremely low.
Conclusion
This pilot study strongly supports the feasibility of studies that combine the use of traditional epidemiological exposure and outcome data from population-based registries with the de novo collection of genetic information from true, population-based cases and controls. This model will facilitate rigorous studies of gene–environment interactions that are particularly important for conditions such as pediatric CD and other complex chronic conditions for which there are no medical or surgical cures, making prevention of paramount importance. These future studies will fill a critical unmet need, as prior epidemiological studies of IBD have not included measures of genetic risk and the large-scale genetic studies have not included simultaneous measures of environmental and/or pharmacological exposures.
Acknowledgment
We thank Dr Jason Luo, PhD, and the Mammalian Genotyping Core at the University of North Carolina at Chapel Hill for performing genotyping associated with this project.
Disclosure
The authors report no conflicts of interest in this work.
References
Jacobsen BA, Fallingborg J, Rasmussen HH, et al. Increase in incidence and prevalence of inflammatory bowel disease in northern Denmark: a population-based study, 1978-2002. Eur J Gastroenterol Hepatol. 2006;18(6):601–606. | ||
Loftus EV Jr, Schoenfeld P, Sandborn WJ. The epidemiology and natural history of Crohn’s disease in population-based patient cohorts from North America: a systematic review. Aliment Pharmacol Ther. 2002;16(1):51–60. | ||
Vernier-Massouille G, Balde M, Salleron J, et al. Natural history of pediatric Crohn’s disease: a population-based cohort study. Gastroenterology. 2008;135(4):1106–1113. | ||
Van Limbergen J, Russell RK, Drummond HE, et al. Definition of phenotypic characteristics of childhood-onset inflammatory bowel disease. Gastroenterology. 2008;135(4):1114–1122. | ||
Longobardi T, Jacobs P, Bernstein CN. Work losses related to inflammatory bowel disease in the United States: results from the National Health Interview Survey. Am J Gastroenterol. 2003;98(5):1064–1072. | ||
Ferguson A, Sedgwick DM, Drummond J. Morbidity of juvenile onset inflammatory bowel disease: effects on education and employment in early adult life. Gut. 1994;35(5):665–668. | ||
Cohen RD. The quality of life in patients with Crohn’s disease. Aliment Pharmacol Ther. 2002;16(9):1603–1609. | ||
Akobeng AK, Suresh-Babu MV, Firth D, Miller V, Mir P, Thomas AG. Quality of life in children with Crohn’s disease: a pilot study. J Pediatr Gastroenterol Nutr. 1999;28(4):S37–S39. | ||
Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134(2):577–594. | ||
Orholm M, Binder V, Sorensen TI, Rasmussen LP, Kyvik KO. Concordance of inflammatory bowel disease among Danish twins. Results of a nationwide study. Scand J Gastroenterol. 2000;35(10):1075–1081. | ||
Halfvarson J, Bodin L, Tysk C, Lindberg E, Jarnerot G. Inflammatory bowel disease in a Swedish twin cohort: a long-term follow-up of concordance and clinical characteristics. Gastroenterology. 2003;124(7):1767–1773. | ||
Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491(7422):119–124. | ||
Loftus EV Jr. Clinical epidemiology of inflammatory bowel disease: incidence, prevalence, and environmental influences. Gastroenterology. 2004;126(6):1504–1517. | ||
Kugathasan S, Amre D. Inflammatory bowel disease – environmental modification and genetic determinants. Pediatr Clin North Am. 2006;53(4):727–749. | ||
Frank L. Epidemiology. When an entire country is a cohort. Science. 2000;287(5462):2398–2399. | ||
Frank L. Epidemiology. The epidemiologist’s dream: Denmark. Science. 2003;301(5630):163. | ||
Schmidt M, Schmidt SA, Sandegaard JL, Ehrenstein V, Pedersen L, Sorensen HT. The Danish National Patient Registry: a review of content, data quality, and research potential. Clin Epidemiol. 2015;7:449–490. | ||
IBD Working Group of the European Society for Paediatric Gastroenterology, Hepatology and Nutrition. Inflammatory bowel disease in children and adolescents: recommendations for diagnosis – the Porto criteria. J Pediatr Gastroenterol Nutr. 2005;41(1):1–7. | ||
Nunes AP, Oliveira IO, Santos BR, et al. Quality of DNA extracted from saliva samples collected with the Oragene DNA self-collection kit. BMC Med Res Methodol. 2012;12:65. | ||
Yang SK, Hong M, Zhao W, et al. Genome-wide association study of Crohn’s disease in Koreans revealed three new susceptibility loci and common attributes of genetic susceptibility across ethnic populations. Gut. 2014;63(1):80–87. | ||
Granlund A, Flatberg A, Ostvik AE, et al. Whole genome gene expression meta-analysis of inflammatory bowel disease colon mucosa demonstrates lack of major differences between Crohn’s disease and ulcerative colitis. PLoS One. 2013;8(2):e56818. | ||
Sarlos P, Kovesdi E, Magyari L, et al. Genetic update on inflammatory factors in ulcerative colitis: review of the current literature. World J Gastrointest Pathophysiol. 2014;5(3):304–321. | ||
Ferguson LR, Huebner C, Petermann I, et al. Single nucleotide polymorphism in the tumor necrosis factor-alpha gene affects inflammatory bowel diseases risk. World J Gastroenterol. 2008;14(29):4652–4661. | ||
Brinar M, Cukovic-Cavka S, Bozina N, et al. MDR1 polymorphisms are associated with inflammatory bowel disease in a cohort of Croatian IBD patients. BMC Gastroenterol. 2013;13:57. | ||
Ho GT, Nimmo ER, Tenesa A, et al. Allelic variations of the multidrug resistance gene determine susceptibility and disease behavior in ulcerative colitis. Gastroenterology. 2005;128(2):288–296. | ||
Hugot JP, Chamaillard M, Zouali H, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411(6837):599–603. | ||
Sugimura K, Taylor KD, Lin YC, et al. A novel NOD2/CARD15 haplotype conferring risk for Crohn disease in Ashkenazi Jews. Am J Hum Genet. 2003;72(3):509–518. | ||
Ningappa M, Higgs BW, Weeks DE, et al. NOD2 gene polymorphism rs2066844 associates with need for combined liver-intestine transplantation in children with short-gut syndrome. Am J Gastroenterol. 2011;106(1):157–165. | ||
Hansen TV, Simonsen MK, Nielsen FC, Hundrup YA. Collection of blood, saliva, and buccal cell samples in a pilot study on the Danish nurse cohort: comparison of the response rate and quality of genomic DNA. Cancer Epidemiol Biomarkers Prev. 2007;16(10):2072–2076. | ||
Randell RL, Gulati AS, Cook SF, et al. Collecting biospecimens from an internet-based prospective cohort study of inflammatory bowel disease (CCFA Partners): a feasibility study. JMIR Res Protoc. 2016;5(1):e3. | ||
Long MD, Cadigan RJ, Cook SF, et al. Perceptions of patients with inflammatory bowel diseases on biobanking. Inflamm Bowel Dis. 2015;21(1):132–138. | ||
Bernstein L. Control recruitment in population-based case-control studies. Epidemiology. 2006;17(3):255–257. | ||
Hazen RJ. Population based control selection and non-response among case-control studies in cancer research. Ann Epidemiol. 2005;15(8):640. | ||
Castaño-Vinyals G, Nieuwenhuijsen MJ, Moreno V, et al. Participation rates in the selection of population controls in a case-control study of colorectal cancer using two recruitment methods. Gac Sanit. 2011;25:353–356. |
Supplementary material
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.