Back to Journals » Cancer Management and Research » Volume 10
Expressions and prognostic values of the E2F transcription factors in human breast carcinoma
Authors Liu ZL, Bi XW, Liu PP, Lei DX, Wang Y, Li ZM, Jiang WQ, Xia Y
Received 26 April 2018
Accepted for publication 21 June 2018
Published 13 September 2018 Volume 2018:10 Pages 3521—3532
DOI https://doi.org/10.2147/CMAR.S172332
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Harikrishna Nakshatri
Ze-Long Liu,* Xi-Wen Bi,* Pan-Pan Liu,* De-Xin Lei, Yu Wang, Zhi-Ming Li, Wen-Qi Jiang, Yi Xia
Department of Medical Oncology, Sun Yat-sen University Cancer Center, State Key Laboratory of Oncology in South China, Collaborative Innovation Centre of Cancer Medicine, Guangzhou, People’s Republic of China
*These authors contributed equally to this work
Abstract: E2F transcription factors (E2Fs) are a family of transcription factors involved in cell proliferation, differentiation, and apoptosis. Their important roles in the development and metastasis of breast carcinoma (BC) have been discovered by previous in vitro and in vivo studies. Yet, expressions and distinct prognostic values of these eight E2Fs in human BC remain unclear in many respects. In this study, we aimed to reveal their roles in BC through analyzing the transcription and survival data of the E2Fs in BC patients from four online databases including ONCOMINE, Breast Cancer Gene-Expression Miner v4.1, cBioPortal for Cancer Genomics, and Kaplan–Meier Plotter. We found the overexpression of E2Fs in BC tissues compared with normal breast tissues, except for E2F4. Higher expression levels of E2Fs, except for E2F4 and E2F6, were associated with higher levels of Scarff–Bloom–Richardson grade of BC. Alterations of E2Fs were found to be significantly correlated with poorer overall survival of BC patients. Through plotting the survival curve in the Kaplan–Meier Plotter, it was found that higher mRNA levels of E2F1, E2F3, E2F7, and E2F8 were associated with poorer relapse-free survival in all BC patients, indicating that they are potential targets for individualized treatments of BC patients. Conversely, higher mRNA expression level of E2F4 predicted better RFS in BC patients, suggesting E2F4 as a new biomarker for BC prognosis. Considering currently available limited evidence, further studies need to be performed to investigate the roles of E2Fs in BC.
Keywords: E2Fs, breast carcinoma, expressions, prognostic values, Kaplan–Meier plot
Introduction
The E2F transcription factors (E2Fs), which were discovered almost 30 years ago, have been confirmed to play significant roles in cell proliferation, differentiation, and apoptosis.1 It is known that there are eight E2F family member genes so far, named E2F1–E2F8 in the order of discovery. The E2Fs came to the forefront of cancer research when they were found to be associated with and regulated by the RB protein, the product of gene mutation in retinoblastoma.2 Cancer-related proliferative roles of E2Fs have been found in many kinds of human cancer, including breast carcinoma (BC). It was found that they regulated tumor development and metastasis in animal models of BC.2,3 These eight E2Fs, however, are supposed to have some specific functions and overlapping roles according to current studies.4
BC, the most common malignant tumor among women in both developed and developing countries, remains one of the leading causes of cancer death among women worldwide.5 BC is supposed to have diverse characteristics in pathology and molecular biology. BC subtypes defined by immunohistochemical expression of estrogen receptor (ER), progesterone receptor (PR), and HER2 provide prognostic values of BC patients.6 In this molecular classification system, triple-negative BC (TNBC) has the worst overall survival (OS) and disease-free survival (DFS),6 and the E2Fs have been implicated in regulation of TNBC.7 Complex genetic mechanisms regulate and control the cell cycle in cancers, including amplification, mutation, and overexpression of the genes encoding the core components in the cell cycle.8 These components include the cyclins, cyclin-dependent kinases (CDKs), CDK inhibitors, and RB1, all of which contribute to activation of the downstream E2Fs, and activation of E2Fs in turn causes unrestricted cell proliferation and divisions.8 Mutations of the RB1 gene or components regulating the CDK-RB-E2F pathway have been identified in nearly all human malignant tumors, including BC.8
The E2Fs, as mentioned before, are supposed to have complex and distinct roles in human BC. Several reports have discovered that amplification of the E2F1 or E2F3 gene locus and overexpression of E2F1 or E2F3 were frequent genetic events in many human malignancies, whereas large chromosomal deletions of regions including the E2F1, E2F2 or E2F3 genes have been detected in some cases.9 The conclusions of current studies on the role of E2F4 are controversial regarding whether it was a suppressor or an activator of carcinogenesis. E2F4 seemed to be able to function as a tumor suppressor or an oncogene through regulating alternative sets of genes in different tissues.9 An increased gene copy number of E2F5 was detected in two independent cohorts of BC patients.10,11 Evidence from several studies found that E2F6 negatively regulated BRCA1 in human cancer cells, functioning as a repressive transcription factor in a histone methyltransferase independent manner on target promoters.12,13 For BC patients receiving tamoxifen treatment, high expression level of E2F7 was associated with elevated risk of relapse and poor prognosis.14 Up-regulation of E2F8 was reported to promote cell proliferation and tumorigenicity in BC by modulating the G1/S phase transition.15 However, due to limited studies at present, the expression patterns, functions, and prognostic values of the E2Fs in human BC have not been clearly elucidated.
Microarray technology, which has developed rapidly during the past few years, has revolutionized DNA and RNA research and has become essential technology for biomedical research.16 Based on comprehensive analysis of gene expression data and survival data published online, we performed this study to clarify and determine the distinct patterns of expression and significance for survival prognosis of eight E2Fs in BC patients.
Materials and methods
Ethics statement
This study was conducted in accordance with the principles of the Declaration of Helsinki, and with the approval from the academic committee of Sun Yat-sen University Cancer Center. All data were obtained from published online research, which undoubtedly contained informed consent.
ONCOMINE
ONCOMINE, a cancer microarray database and web-based data-mining platform,17 was applied to analyze the mRNA levels of E2Fs in BC. We searched ONCOMINE (www.oncomine.org) for the fold changes of E2Fs in BC using the filters of differential analysis (cancer vs normal), cancer type (breast cancer), sample type (clinical specimen), data type (mRNA), and gene (E2F1, E2F2, E2F3, E2F4, E2F5, E2F6, E2F7, or E2F8). The comparisons of mRNA levels of E2Fs in BC and normal tissues in each individual dataset were conducted using the Student’s t-test. We then conducted the meta-analysis of differential expression of E2Fs in BC vs normal tissues. Random-effects models were employed in the meta-analysis according to the method previously described elsewhere.18
The Breast Cancer Gene-Expression Miner v4.1
The Breast Cancer Gene-Expression Miner v4.1 (bcGenExMiner v4.1) is a statistical mining tool of 36 published annotated genomic datasets (total of 5,861 patients) and has three statistical analysis functions, as listed in the following paragraphs.19,20 The expression module permitted comparisons of expressions of candidate genes according to several clinical criteria, such as age, nodal status, ER status, PR status, HER2 status, and so on. The prognostic module evaluated the prognostic values of candidate genes in human BC and the correlation module permitted analysis of the correlations between candidate genes.
The cBioPortal for Cancer Genomics
The cBioPortal for Cancer Genomics provides visualization, analysis, and downloading of large-scale cancer genomics datasets.21,22 The breast cancer dataset (METABRIC, Nature 2012),23 which contains data, including histopathological data of 2,509 BC patients, was chosen for analyses of E2Fs using the cBioPortal for Cancer Genomics (www.cbioportal.org). Selected genomic profiles included mutations, copy-number variance from GISTIC, and mRNA expression z scores (Illumina Human v3 microarray) with a z score threshold of ±2.0. OS was calculated with the Kaplan–Meier survival curve according to the instruction on the website.
The Kaplan–Meier Plotter
The Kaplan–Meier Plotter (www.cbioportal.org),24 which collected miRNA expression data and survival data of a total of 5,143 BC patients from gene expression omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/), The Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov/), European Genome-phenome Archive (EGA) (https://ega.crg.eu/), and PubMed (http://www.pubmed.com),25,26 was used to explore the prognostic values of mRNA levels of E2Fs in BC. BC patients were divided into two groups by the median mRNA expression level (high expression vs low expression) in order to analyze the OS and relapse-free survival (RFS) with Kaplan–Meier plots, in which the number-at-risk was listed. Only the JetSet best probe set of E2Fs was selected for our analysis.27 The HR with 95% CI and the log-rank P-value was calculated in each Kaplan–Meier survival plot and the cutoff of log-rank P-value was defined as 0.05.
Results
The transcription levels of E2Fs in BC compared with that in normal tissues
A total of 13 datasets containing 3,555 samples were included in this study, of which the largest two are the Curtis dataset (2,136 samples) and TCGA dataset (593 samples). The Curtis dataset collected breast cancer specimens from tumor banks in the UK and Canada,28 while TCGA dataset was generated by the National Cancer Institute (NCI) in the USA. Through conducting meta-analysis based on the ONCOMINE databases, we compared the transcription levels of eight E2Fs in BC and normal tissues (Tables S1–S8) We found that the mRNA expression levels of E2F1, E2F2, E2F3, E2F5, E2F6, E2F7, and E2F8 were significantly higher in BC tissues, with fold changes of 1.63, 2.07, 1.53, 1.61, 1.21, 2.27, and 2.05, respectively (Table 1). However, there was no significant difference between the transcription levels of E2F4 in BC and normal tissues (fold change =1.13, 95% CI: 0.87–1.40).
The mRNA levels of E2Fs are correlated with clinical and molecular features of BC patients
The Welch’s tests, along with Dunnett-Tukey-Kramer’s tests for pairwise comparison when appropriate, were performed to compare the mRNA levels of E2Fs between groups of patients divided according to different clinical and molecular criteria in the bcGenExMiner v4.1. For the criterion of age, it was found that no significant difference existed between ≤51 years old and >51 years old groups of E2F1, E2F4, E2F7, and E2F8, whereas downregulated expression of E2F2, E2F3, E2F5, and E2F6 in the older group was found (Table 2). BC patients with positive nodal status showed higher mRNA level of E2F5 than negative nodal patients (Table 2).
Higher Scarff Bloom & Richardson (SBR) grade status was found to be correlated with higher mRNA levels of all E2Fs (Figure 1). For E2F4 and E2F6, although a significant difference was detected in the Welch’s test, the group comparison between SBR1 and SBR2 by Dunnett-Tukey-Kramer’s test of both did not show a significant difference (the cutoff value of P is 0.05) (Table S9).
We found that ER status was negatively associated with mRNA levels of E2Fs except for E2F6, whereas PR status was negatively associated with mRNA levels of E2Fs except for E2F5. In HER2-positive groups of BC patients, the transcription levels of E2F1, E2F3, E2F7, and E2F8 were significantly up-regulated compared with HER2-negative groups. As mentioned before, TNBC is a special type of BC with negative ER, PR, and HER2, and has the worst clinical outcome. The mRNA levels of E2Fs, except for E2F4 and E2F6, were found to be significantly higher in TNBC patients (Table 2).
BC patients with alterations of E2Fs have poorer OS
Among the overall 2,509 patients with breast invasive carcinoma in the selected dataset, 1,120 (44.6%) were detected to have alterations of E2Fs (Figure 2A). E2F5 was altered in 22% of BC patients in this dataset. BC patients with alterations of E2Fs were found to have significantly poorer OS according to analyses by log-rank tests in the Kaplan–Meier survival plots (P=0.0249) (Figure 2B).
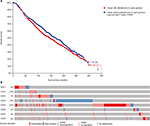  | Figure 2 Analysis of E2Fs’ alterations in breast invasive carcinoma (using cBioPortal for Cancer Genomics21,22). Notes: (A) The Kaplan–Meier plot comparing overall survival of breast carcinoma patients with E2F alterations (n=1,120) and without E2F alternations (n=1,389). (B) Oncoprint in cBioPortal represented the proportion and distribution of cases with E2F alterations. The figure was cropped on the right to exclude cases without alterations. Abbreviation: E2Fs, E2F transcription factors. |
Higher mRNA levels of E2F1, E2F3, and E2F8 were associated with poorer OS and RFS of BC patients
It was found that mRNA levels of E2F1, E2F3, and E2F8 were significantly correlated with OS and RFS in all BC patients (P<0.05) (Figures 3 and 4), through analyses by log-rank tests in the Kaplan–Meier survival plots. Higher mRNA levels of E2F1, E2F3, and E2F8 predicted poorer OS and RFS in BC patients. In contrast, BC patients with higher mRNA levels of E2F4 were found to have better RFS. Additionally, transcription levels of E2F7 were negatively associated with RFS but not OS in BC patients.
Discussion
The E2Fs were involved in BC development and metastasis, and demonstrated prognostic values according to currently available limited studies. However, the multifaceted roles of E2Fs in the development, metastasis, and prognostication of BC remain to be clarified. As far as we know, this is the first study that systematically analyzed the mRNA expression levels and prognostic values of the eight E2Fs in human BC.
E2F1, the first member of the family of E2Fs, was proven to initiate and maintain tumors originating from distinct tissues in multiple mouse models.29 However, some studies demonstrated that they induced cell apoptosis and resulted in inhibition of tumor growth in some specific tissue types, such as the skin.29 As for BC, Wu et al found that E2F1 played an oncogenic role in ErbB2- or Myc-triggered mammary tumorigenesis.30 Moreover, the low transcription level of E2F1 was reported as a strong determinant of favorable outcome for BC patients.31 In this study, we found that the mRNA level of E2F1 was significantly up-regulated in BC. Higher mRNA level of E2F1 was associated with higher SBR grade and TNBC, which predicted higher degree of malignancy, higher incidence of recurrence and metastasis, and worse clinical outcomes. In survival analysis, BC patients with higher mRNA level of E2F1 were found to have poorer OS and RFS. We thus suppose E2F1 as a target for precision therapy of BC patients, and a previous study in cell lines found that MIR372 inhibited proliferation and induced apoptosis in BC cells by directly targeting E2F1.32
Significant reductions in tumor incidence, the metastatic capacity of the tumor and the number of circulating tumor cells in animals with E2F2 knockout background were found in numerous studies.2,33 E2F2 loss resulted in increased metastasis of BC, potentially functioning through a PTPRD-dependent mechanism.22 Interestingly, on the contrary, Wu et al noted a tumor suppressor role of E2F2 in Myc-mediated mammary tumorigenesis.30 The mRNA level of E2F2 was found to be significantly higher in BC, especially in TNBC, and it was positively associated with the SBR grade as E2F1. However, unlike E2F1, mRNA expression level of E2F2 did not have prognostic values for OS or RFS of BC patients. Nguyen-Vu et al reported that LXR ligand treatment downregulated transcription level of E2F2 and resulted in significant disruption of cell proliferation in ER-positive BC.34 This finding supported that E2F2 might also be a potential treatment target for BC. Considering the small number of studies focused on functions of E2F in BC, more work needs to be carried out in future.
The oncogenic activity of E2F3 has been observed in ErbB2- or Myc-triggered mammary tumorigenesis.30 Fujiwara et al noted a significant reduction in tumor incidence with the loss of E2F3,33 whereas Lee et al found that E2F3 silencing inhibited mammary tumor growth through reducing the percentage of cells undergoing mitosis.35 An in vitro study demonstrated that E2F3 was a diagnostic and potential therapeutic target in BC.36 In this study, the mRNA level of E2F3 was found to be significantly higher in BC. Higher mRNA level of E2F3 was associated with higher SBR grade and TNBC. Survival analysis revealed that higher mRNA levels of E2F3 predicted poorer OS and RFS in BC patients, and we thus suppose E2F3 as another therapeutic target for BC patients. A previous study found that metformin reduced the incidence of breast cancer and metastasis by increasing miR-26a expression, which downregulated the expression level of E2F3.37 Another in vitro study reported that T-VISA-miR-34a induced expression of miR-34a, and dramatically suppressed growth, migration, and invasion of breast cancer cells by downregulating the protein expression levels of target genes including E2F3.38 Vimala et al found that siRNA for E2F3 facilitated the silencing of E2F3 overexpression and “fought against” BC in cell lines.36 Results of these studies are consistent with our assumption.
It was reported that E2F4 had an oncogenic role rather than a tumor suppressor role in breast carcinogenesis, and expression of E2F4 in invasive BC was associated with poor prognosis.39 Although there was no significant up-regulation or downregulation of E2F4 expression level in BC, BC patients with higher mRNA levels of E2F4 were found to have significantly better RFS. We thus supposed that E2F4 was a potential prognostic marker for better survival of BC patients. Considering the currently available little evidence and the unavoidable limitations of this study, further research needs to be performed to investigate the role of E2F4 in BC.
Polanowska et al found that E2F5 was oncogenic in primary rodent cells and was amplified in human BC,10 whereas Umemura et al found that E2F5-positive subtype of BC was associated with a basal phenotype, TNBC, and worse clinical outcome.11 In this study, it was found that the mRNA level of E2F5 was significantly higher in BC. Higher mRNA level of E2F5 was correlated with higher SBR grade and TNBC. BC patients with positive nodal status showed significantly higher mRNA level of E2F5 than negative nodal patients. This finding supported that E2F5 might play a role in lymph node metastasis of BC patients. Through targeting E2F5 with miR-154 in cell lines, the growth and invasion of breast cancer cells were inhibited.40 However, mRNA expression levels of E2F5 did not have prognostic values in BC patients according to our study.
Several current studies found that E2F6 negatively regulated BRCA1 in BC.12,13 The mRNA level of E2F6 was found to be significantly higher in BC, and higher mRNA level was found in higher SBR grade of BC patients, which indicated worse clinical outcomes. However, no significant difference of OS or RFS was found between BC patients with high and low mRNA level of E2F6. As little research has focused on E2F6 so far, the underlying role of E2F6 in BC needs more investigation.
E2F7 and E2F8 were both supposed to promote tumorigenicity in breast cancer according to currently available limited studies.14,15 We found that both of their transcription levels were significantly higher in BC. BC patients with higher SBR grade and TNBC patients had higher mRNA levels of E2F7 and E2F8. Not surprisingly, the transcription levels of E2F7 and E2F8 were negatively associated with RFS of BC patients. Higher E2F8 expression level also predicted worse OS of BC patients. These results indicate that E2F7 and E2F8 are both potential new targets for individualized treatments of BC patients, and further studies need to be performed to explore their potential values.
We also found that alterations of E2Fs were frequent genetic events in BC patients and BC patients with alterations of E2Fs appeared to have significantly poorer OS, although the underlying mechanism is still unclear.
Conclusion
We performed comprehensive analyses on the expressions and prognostic values of the eight E2Fs in BC in this study, for the first time, to provide a better understanding of the diversity of BC regarding various aspects including clinical, histopathological, and biomolecular characteristics. Our results indicate that E2F1, E2F3, E2F7, and E2F8 are potential targets for individualized treatment of BC patients, whereas E2F4 is a potential prognostic marker for better survival of BC patients.
Acknowledgment
This work was supported by the National Natural Science Foundation of China (nos. 81071950, 81301903).
Disclosure
The authors report no conflicts of interest in this work.
References
Degregori J, Johnson DG. Distinct and Overlapping Roles for E2F Family Members in Transcription, Proliferation and Apoptosis. Curr Mol Med. 2006;6(7):739–748. | ||
Hollern DP, Honeysett J, Cardiff RD, Andrechek ER. The E2F transcription factors regulate tumor development and metastasis in a mouse model of metastatic breast cancer. Mol Cell Biol. 2014;34(17):3229–3243. | ||
Andrechek ER. HER2/Neu tumorigenesis and metastasis is regulated by E2F activator transcription factors. Oncogene. 2015;34(2):217–225. | ||
Attwooll C, Lazzerini Denchi E, Helin K. The E2F family: specific functions and overlapping interests. Embo J. 2004;23(24):4709–4716. | ||
Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. | ||
Onitilo AA, Engel JM, Greenlee RT, Mukesh BN. Breast cancer subtypes based on ER/PR and Her2 expression: comparison of clinicopathologic features and survival. Clin Med Res. 2009;7(1–2):4–13. | ||
Verlinden L, vanden Bempt I, Eelen G, et al. The E2F-regulated gene Chk1 is highly expressed in triple-negative estrogen receptor /progesterone receptor /HER-2 breast carcinomas. Cancer Res. 2007;67(14):6574–6581. | ||
Johnson J, Thijssen B, Mcdermott U, et al. Targeting the RB-E2F pathway in breast cancer. Oncogene. 2016;35(37):4829–4835. | ||
Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer. 2009;9(11):785–797. | ||
Polanowska J, Le Cam L, Orsetti B, et al. Human E2F5 gene is oncogenic in primary rodent cells and is amplified in human breast tumors. Genes Chromosomes Cancer. 2000;28(1):126–130. | ||
Umemura S, Shirane M, Takekoshi S, et al. Overexpression of E2F-5 correlates with a pathological basal phenotype and a worse clinical outcome. Br J Cancer. 2009;100(5):764–771. | ||
Oberley MJ, Inman DR, Farnham PJ. E2F6 negatively regulates BRCA1 in human cancer cells without methylation of histone H3 on lysine 9. J Biol Chem. 2003;278(43):42466–42476. | ||
Yang WW, Wang ZH, Zhu Y, Yang HT. E2F6 negatively regulates ultraviolet-induced apoptosis via modulation of BRCA1. Cell Death Differ. 2007;14(4):807–817. | ||
Chu J, Zhu Y, Liu Y, et al. E2F7 overexpression leads to tamoxifen resistance in breast cancer cells by competing with E2F1 at miR-15a/16 promoter. Oncotarget. 2015;6(31):31944–31957. | ||
Ye L, Guo L, He Z, et al. Upregulation of E2F8 promotes cell proliferation and tumorigenicity in breast cancer by modulating G1/S phase transition. Oncotarget. 2016;7(17):23757. | ||
Sealfon SC, Chu TT. RNA and DNA microarrays. Methods Mol Biol. 2011;671(671):3–34. | ||
Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. | ||
Rest JS, Wilkins O, Yuan W, Purugganan MD, Gurevitch J. Meta-analysis and meta-regression of transcriptomic responses to water stress in Arabidopsis. Plant J. 2016;85(4):548–560. | ||
Jézéquel P, Campone M, Gouraud W, et al. bc-GenExMiner: an easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res Treat. 2012;131(3):765–775. | ||
Jézéquel P, Frénel JS, Campion L, et al. bc-GenExMiner 3.0: new mining module computes breast cancer gene expression correlation analyses. Database (Oxford). 2013;2013:bas060. | ||
Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. | ||
Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. | ||
Pereira B, Chin SF, Rueda OM, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7:11479. | ||
Győrffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8(12):e82241. | ||
Györffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123(3):725–731. | ||
Lánczky A, Nagy Á, Bottai G, et al. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat. 2016;160(3):439–446. | ||
Li Q, Birkbak NJ, Gyorffy B, Szallasi Z, Eklund AC, et al. Jetset: selecting the optimal microarray probe set to represent a gene. BMC Bioinformatics. 2011;12(1):474–477. | ||
Curtis C, Shah SP, Chin SF, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. | ||
Lee M, Rivera-Rivera Y, Moreno CS, Saavedra HI. The E2F activators control multiple mitotic regulators and maintain genomic integrity through Sgo1 and BubR1. Oncotarget. 2017;8(44):77649. | ||
Wu L, de Bruin A, Wang H, et al. Selective roles of E2Fs for ErbB2- and Myc-mediated mammary tumorigenesis. Oncogene. 2015;34(1):119–128. | ||
Vuaroqueaux V, Urban P, Labuhn M, et al. Low E2F1 transcript levels are a strong determinant of favorable breast cancer outcome. Breast Cancer Res. 2007;9(3):R33. | ||
Zhao YX, Liu HC, Ying WY, et al. microRNA372 inhibits proliferation and induces apoptosis in human breast cancer cells by directly targeting E2F1. Mol Med Rep. 2017;16(6):8069–8075. | ||
Fujiwara K, Yuwanita I, Hollern DP, et al. Prediction and genetic demonstration of a role for activator E2Fs in Myc-induced tumors. Cancer Res. 2011;71(5):1924–1932. | ||
Nguyen-Vu T, Vedin LL, Liu K, et al. Liver × receptor ligands disrupt breast cancer cell proliferation through an E2F-mediated mechanism. Breast Cancer Res. 2013;15(3):R51. | ||
Lee M, Oprea-Ilies G, Saavedra HI. Silencing of E2F3 suppresses tumor growth of Her2+ breast cancer cells by restricting mitosis. Oncotarget. 2015;6(35):37316–37334. | ||
Vimala K, Sundarraj S, Sujitha MV, Kannan S. Curtailing overexpression of E2F3 in breast cancer using siRNA (E2F3)-based gene silencing. Arch Med Res. 2012;43(6):415–422. | ||
Cabello P, Pineda B, Tormo E, Lluch A, Eroles P. The Antitumor Effect of Metformin Is Mediated by miR-26a in Breast Cancer. Int J Mol Sci. 2016;17(8):1298. | ||
Li L, Xie X, Luo J, et al. Targeted expression of miR-34a using the T-VISA system suppresses breast cancer cell growth and invasion. Mol Ther. 2012;20(12):2326–2334. | ||
Rakha EA, Pinder SE, Paish EC, Robertson JF, Ellis IO. Expression of E2F-4 in invasive breast carcinomas is associated with poor prognosis. J Pathol. 2004;203(3):754–761. | ||
Xu H, Fei D, Zong S, Fan Z. MicroRNA-154 inhibits growth and invasion of breast cancer cells through targeting E2F5. Am J Transl Res. 2016;8(6):2620–2630. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.