Back to Journals » OncoTargets and Therapy » Volume 11
Expression profiles of long noncoding RNAs in lung adenocarcinoma
Authors Zhao B , Xu H, Ai X, Adalat Y, Tong Y, Zhang J, Yang S
Received 8 March 2018
Accepted for publication 13 May 2018
Published 6 September 2018 Volume 2018:11 Pages 5383—5390
DOI https://doi.org/10.2147/OTT.S167633
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Leo Jen-Liang Su
Bing Zhao,1 Hang Xu,2 Xiaoye Ai,1 Yasheng Adalat,1 Ying Tong,1 Jiazhennan Zhang,1 Shune Yang3
1Department of Day Oncology, Affiliated Tumor Hospital of Xinjiang Medical University, Urumqi, People’s Republic of China; 2Intensive Care Unit, The Sixth Affiliated Hospital, Xinjiang Medical University, Urumqi, Xinjiang, People’s Republic of China; 3Department of Breast Cancer and Lymphoma, Affiliated Tumor Hospital of Xinjiang Medical University, Urumqi, People’s Republic of China
Purpose: This study aimed to analyze expression profiles of long noncoding RNAs (lncRNAs) in lung adenocarcinoma.
Methods: lncRNA microarray technology was employed to detect lncRNA profiles of 3 pairs of lung adenocarcinoma tissues and adjacent tissues.
Results: We found 134 upregulated lncRNAs and 460 downregulated lncRNAs in lung adenocarcinoma tissues compared to adjacent tissues. Among them, LINC00152, LINC00691, and LINC00578 showed the most significant changes of upregulation, while LINC00668, LINC00710, and LINC00607 showed the most significant changes of downregulation. Fluorescent quantitative polymerase chain reaction (PCR) analysis of tissue samples from an additional 90 patients with lung adenocarcinoma showed significantly increased levels of LINC00152, LINC00691, and LINC00578 and decreased levels of LINC00668, LINC00710, and LINC00607 in lung adenocarcinoma tissues. In addition, LINC00578 was closely associated with the existence of metastasis of lung adenocarcinoma, but the other 5 lncRNAs showed no significant correlation with clinicopathologic characteristics such as age, gender, tumor stage, and the existence of metastasis. Further follow-up study showed that LINC00578 expression was closely associated with the survival of patients with lung adenocarcinoma.
Conclusion: We revealed the expression profiles of lncRNAs in lung adenocarcinoma and identified LINC00578 as a promising biomarker and therapeutic target for lung adenocarcinoma.
Keywords: long noncoding RNA, microarray, lung adenocarcinoma, survival
Introduction
Lung cancer is one of the malignant tumors with the biggest threat to human health and life. In the last 50 years, there has been significant increase in the morbidity and mortality of lung cancer worldwide. The morbidity and mortality of male lung cancer rank at the top of the list all malignant tumors, while the morbidity and mortality of female lung cancer rank the second. About 80%–90% of patients are at advanced stage and have distant metastasis when lung cancer is detected, and thus are not suitable for radical surgeries.1–3 Among pathological types, lung adenocarcinoma accounts for 40% of all non-small-cell lung cancers, and it metastasizes at an early stage, has poor prognosis, and has low treatment efficacy.2,3 Therefore, it is particularly important to elucidate molecular mechanisms for the metastasis of lung adenocarcinoma in order to develop novel approaches to early diagnosis and effective treatment of lung adenocarcinoma.
Long noncoding RNAs (lncRNAs) are noncoding RNAs <200 nucleotides long. lncRNAs play important role in dosage compensation, epigenetic control, cell cycle control, and cell differentiation.4–6 The abnormal expression or function of lncRNAs is closely related to the occurrence of human diseases such as cancer and neurodegenerative disease.7–10 However, current literature on the role of lncRNAs in malignant lung adenocarcinoma is rare. This study aimed to compare lncRNA profiles between lung adenocarcinoma tissues and paired adjacent tissues using lncRNA microarray technology and identify lncRNAs associated with the occurrence of lung adenocarcinoma as novel biomarkers and therapeutic targets for lung adenocarcinoma.
Materials and methods
Patients and tissues
All patients gave written informed consent and the study was approved by Ethics Committee of Tumor Hospital Affiliated to Xinjiang Medical University. Lung adenocarcinoma tissues and normal adjacent tissues were dissected from 3 patients who underwent lung adenocarcinoma surgery from July 2012 to June 2015 for microarray analysis, and lung adenocarcinoma was pathologically verified. Patients with advanced lung adenocarcinoma with the complication of liver metastasis, brain metastasis, bone metastasis, and abdominal cavity metastasis were excluded because these conditions affected their survival and interfered with the analysis of the correlation of lncRNAs with clinicopathologic characteristics. Patients who had previously received radical lung adenocarcinoma surgery, radiotherapy, and chemotherapy were excluded. For verification study, an additional 90 patients with lung adenocarcinoma were recruited and their data are listed in Table 1.
  | Table 1 Clinical data of 90 patients |
Microarray analysis
Microarray of lncRNAs was performed by Beijing CapitalBio Technology Co., Ltd (Beijing, People’s Republic of China). lncRNAs from all canonical databases, NCBI Refseq, UCSC Known Gene, NRED, and RNAdb, were covered with a 12×135 k lncRNA microarray manufactured by Arraystar (Rockville, MD, USA). The probes of microarray were made of 60 mer long oligonucleotides. More than 1 probe was designed for each sequence to increase the reliability of signals.
Total RNA was extracted from tissue samples using Trizol (Invitrogen, Carlsbad, CA, USA) at the ratio of 50 mg sample/mL Trizol. The quality of RNA was analyzed by formaldehyde-denatured gel electrophoresis. Reverse transcription was conducted and ds-cDNA was synthesized using a ds-cDNA synthesis kit (Invitrogen). ds-cDNA was labeled using a monochromatic DNA labeling kit (NimbleGen, Madison, WI, USA). The efficiency of fluorescent labeling was tested using a spectrophotometer. Later, the labeled ds-cDNAs were sequenced and hybridized using a hybridization system (NimbleGen). The fluorescence intensity of microarray was scanned using Axon GenePix Scanner 4000B (Axon Instruments, Union City, CA, USA) and analyzed with NimbleScan software (Ver 2.5) (NimbleGen). GO and pathway analysis was performed using DAVID bioinformatic resources 6.7 (http://david.abcc.ncifcrf.gov) and KEGG Pathway Database (http://www.genome.jp/kegg/pathway.html) to predict lncRNA function and the annotation of coexpressed genes.
Construction of coexpression network
Coexpression analysis of the coding–noncoding genes that included the differentially expressed lncRNAs and nearby coding genes (distance <300 kb) was conducted in R environment using RedeR package as described previously.11 Pearson’s correlation coefficient was not <0.94. The P-values for enrichment between differentially expressed lncRNAs and transcription factors (TFs) was calculated using the hypergeometric distribution method, and the recommended P-value cut-off was 0.05.
Real-time polymerase chain reaction (PCR)
Tumor tissues and adjacent tissues were excised from 90 lung adenocarcinoma patients and resuspended in Trizol (Invitrogen) at the ratio of 50 mg sample/mL Trizol. Tissues were homogenized and total RNA was extracted following the manufacturer’s protocols. Reverse transcription was conducted using Quant Script RT kit (Takara, Kyoto, Japan) according to the user’s manual. Real-time PCR was performed for LINC00578, LINC00668, LINC00710, LINC00607, LINC00152, LINC00691, and GAPDH using SYBR Green Real-time PCR Masker Mix kit (Takara) on 7900HT fluorescent quantitative PCR machine (ABI). Primers are listed in Table 2. GAPDH was used as the internal control, and the fold change was calculated.
  | Table 2 Primers used for real-time polymerase chain reaction (PCR) |
Statistical analysis
Statistical analyses were accomplished using SAS software 9.3 (SAS Institute Inc., Cary, NC, USA), with all data expressed as means ± SD. The correlation between lncRNA and clinical and pathological characteristics was analyzed using Pearson χ2 test. The Student’s t-test was used to evaluate the significance of difference between groups. P<0.05 was considered as significant.
Results
Microarray analysis
Microarray analysis of 3 pairs of qualified samples from lung adenocarcinoma tissues and adjacent tissues showed that a total of 6,960 lncRNAs were detected. After clustering analysis and comparison, 594 lncRNAs were differentially expressed. Among them, 134 lncRNAs were upregulated and 460 lncRNAs were downregulated in tumor tissues compared to adjacent tissues. Notably, 12 lncRNAs had levels increased by >5 times and 121 lncRNAs had levels decreased by >5 times in tumor tissues than in adjacent tissues (Tables 3 and 4).
  | Table 3 Top 10 upregulated lncRNAs with fold change >5 in tumor tissues |
  | Table 4 Top 10 downregulated lncRNAs with fold change >5 in tumor tissues |
To verify microarray results, we focused on 6 lncRNAs which showed the most significant changes of upregulation or downregulation: LINC00152, LINC00691, LINC00578, LINC00668, LINC00710, and LINC00607, and employed quantitative PCR to detect their expression in 90 pairs of lung adenocarcinoma tissues and adjacent tissues. The expression of LINC00152, LINC00691, and LINC00578 was increased significantly, while the expression of LINC00668, LINC00710, and LINC00607 was decreased significantly in lung adenocarcinoma tissues compared to adjacent tissues (Figure 1). Thus, both microarray and PCR analysis showed the upregulated expression of LINC00152, LINC00691, and LINC00578 and the downregulated expression of LINC00668, LINC00710, and LINC00607.
  | Figure 1 Differentially expressed lncRNAs were confirmed by RT-PCR. |
Pearson correlation analysis showed that LINC00578 was closely associated with the metastasis of lung adenocarcinoma (r=0.92). However, there was no significant correlation of the clinicopathologic characteristics such as age, gender, tumor stage, and metastasis with the expression levels of the other 5 lncRNAs (r<0.864) (Table 5).
  | Table 5 The correlation of the expression of the 6 selected lncRNAs with clinicopathological parameters |
Bioinformatics analysis
Using GO cluster analysis, we found that lncRNAs upregulated in lung adenocarcinoma tissues were involved in a variety of malignant phenotypes related to tumorigenesis, such as cell cycle control and mitotic control (Figure 2). In addition, we found several signaling pathways in which upregulated lncRNA are involved (Figure 3).
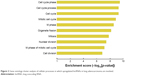  | Figure 2 Gene ontology cluster analysis of cellular processes in which upregulated lncRNAs in lung adenocarcinoma are involved. |
  | Figure 3 Gene ontology cluster analysis of signaling pathways in which upregulated lncRNAs in lung adenocarcinoma are involved. |
Construction of the coexpression network
We constructed a coexpression network composed of 641 nodes and 571 connections between 109 differentially expressed lncRNAs and 532 coding genes. Notably, most of these potential transregulatory lncRNAs could participate in pathways regulated by 3 transcription factors (TFs): NF-κB1, SPI1, and BATF. In the core network of 1,092 lncRNA–TF pairs, NF-κB1 participates in 222 pairs, SPI1 participates in 439 pairs, and BATF participates in 431 pairs (Figure 4). When the top 300 coexpression pairs in P-value order were clustered in lncRNA–TF–mRNA network, NF-κB was identified as the transcription regulator that plays a central role in signaling pathways (Figure 5).
Relationship between LINC00578 expression and patient survival
Of the 90 patients with lung adenocarcinoma, the median follow-up was 34 months (range, 4–60 months), with 52 patients alive and 38 cancer-related deaths at the last clinical follow-up. The median disease-free survival (DFS) and overall survival (OS) were 28 and 32 months, respectively. The 1-, 3-, and 5-year survival rates for the entire cohort of patients were 85.6%, 62.2%, and 13.3%, respectively. By Kaplan–Meier analysis, LINC00578 expression was closely associated with DFS and OS. For the whole cohort, median DFS was longer for patients with low LINC00578 expression than for patients with high LINC00578 expression (34 vs 22 months; P=0.008; Figure 6A). Median OS was also longer for patients with low LINC00578 expression than for patients with high LINC00578 expression (37 vs 25 months; P=0.031; Figure 6B).
Discussion
With the development of genome sequencing technology, it was found that >90% of human genomic DNA can be transcribed into RNAs, of which only 2% encode proteins.12 lncRNAs account for 80% of all noncoding RNAs.13 Different from micro-RNAs (miRNAs), lncRNAs refer to noncoding RNAs with length >200 nt, and they have a specific and complex secondary structure which can provide many loci to bind proteins and interact with DNA and RNA specifically and dynamically through complementary base pairing, thus forming a complex and delicate gene expression control network.14–16
Recent studies have shown that lncRNAs play an important role in tumorigenesis, such as the regulation of invasion and metastasis of malignant tumors.17–20 In particular, a novel lncRNA MUC5B-AS1 was identified to be upregulated in lung adenocarcinoma tissues and promote metastasis of lung adenocarcinoma through regulating MUC5B expression.21 A recent study reported that LINC00707 promoted lung adenocarcinoma cell proliferation and migration through regulating Cdc42 expression.22 Another recent study reported that overexpression of LINC00319 indicated poor prognosis of lung adenocarcinoma patients, and LINC00319 contributed to lung adenocarcinoma progression by modulating miR-450b-5p/EZH2 pathway.23 However, only few studies have analyzed lncRNA profiles in cancers on a large scale, such as high-throughput RNA-seq sequencing.24 Currently, the molecular mechanism by which lncRNAs participate in lung adenocarcinoma remains elusive. Therefore, it is of great significance to explore lncRNAs involved in lung adenocarcinoma to provide a new strategy for treating lung adenocarcinoma.
In this study, we collected 3 pairs of lung adenocarcinoma tissues and adjacent tissues and analyzed the differences in lncRNA profiles. The results showed that 594 lncRNAs changed significantly (fold change >2), of which 134 were upregulated and 460 were downregulated. In particular, the levels of 12 lncRNAs increased by >5 times and the levels of 121 lncRNAs decreased by >5 times, indicating that these lncRNAs are involved in the tumorigenesis of lung adenocarcinoma.
lncRNAs regulate the expression and modification of coding genes in a variety of ways, including epigenetic control, transcriptional control, post-transcriptional control, and cytoplasm transport.25–27 To further validate the microarray results, we performed fluorescent quantitative PCR on 6 selected lncRNAs in samples from additional 90 patients with lung adenocarcinoma. The results confirmed the upregulated and downregulated lncRNAs. In addition, we found that LINC00578 was closely associated with the metastasis of lung adenocarcinoma, but there was no significant correlation of clinicopathologic characteristics with the other 5 lncRNAs. Further follow-up study showed that LINC00578 expression was closely associated with DFS and OS of patients with lung adenocarcinoma. Collectively, these data indicate that LINC00578 is a potential biomarker and therapeutic target for lung adenocarcinoma.
In summary, we explored the expression profiles of lncRNAs in lung adenocarcinoma and identified LINC00578 as a promising biomarker and therapeutic target. Further in-depth studies are needed to elucidate the mechanism of the differentially expressed lncRNAs in lung adenocarcinoma in order to provide novel approaches for the clinical treatment of lung adenocarcinoma.
Acknowledgment
This study was supported by Natural Science Foundation of Xinjiang Uyghur autonomous region (No 2016D01C368).
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. | ||
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. | ||
Rami-Porta R, Asamura H, Travis WD, Rusch VW. Lung cancer – major changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J Clin. 2017;67(2):138–155. | ||
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. | ||
Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. | ||
Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338(6113):1435–1439. | ||
Dey BK, Mueller AC, Dutta A. Long non-coding RNAs as emerging regulators of differentiation, development, and disease. Transcription. 2014;5(4):e944014. | ||
Zhao W, An Y, Liang Y, Xie XW. Role of HOTAIR long noncoding RNA in metastatic progression of lung cancer. Eur Rev Med Pharmacol Sci. 2014;18(13):1930–1936. | ||
Ricciuti B, Mencaroni C, Paglialunga L, et al. Long noncoding RNAs: new insights into non-small cell lung cancer biology, diagnosis and therapy. Med Oncol. 2016;33(2):18. | ||
Tao H, Yang JJ, Zhou X, Deng ZY, Shi KH, Li J. Emerging role of long noncoding RNAs in lung cancer: Current status and future prospects. Respir Med. 2016;110:12–19. | ||
Castro MA, Wang X, Fletcher MN, Meyer KB, Markowetz F. RedeR: R/Bioconductor package for representing modular structures, nested networks and multiple levels of hierarchical associations. Genome Biol. 2012;13(4):R29. | ||
Meseure D, Drak Alsibai K, Nicolas A, Bieche I, Morillon A. Long noncoding RNAs as new architects in cancer epigenetics, prognostic biomarkers, and potential therapeutic targets. Biomed Res Int. 2015;2015:320214. | ||
Tang JY, Lee JC, Chang YT, et al. Long noncoding RNAs-related diseases, cancers, and drugs. ScientificWorldJournal. 2013;2013:943539. | ||
Sen R, Ghosal S, Das S, Balti S, Chakrabarti J. Competing endogenous RNA: the key to posttranscriptional regulation. ScientificWorldJournal. 2014;2014:896206. | ||
Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol. 2010;220(2):126–139. | ||
Hung T, Chang HY. Long noncoding RNA in genome regulation: prospects and mechanism. RNA Biol. 2010;7(5):582–585. | ||
Zhang F, Zhang L, Zhang C. Long noncoding RNAs and tumorigenesis: genetic associations, molecular mechanisms, and therapeutic strategies. Tumour Biol. 2016;37(1):163–175. | ||
Gutschner T, Hämmerle M, Eissmann M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73(3):1180–1189. | ||
Kazemzadeh M, Safaralizadeh R, Orang AV. LncRNAs: emerging players in gene regulation and disease pathogenesis. J Genet. 2015;94(4):771–784. | ||
Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25(10):1335–1337. | ||
Yuan S, Liu Q, Hu Z, et al. Long non-coding RNA MUC5B-AS1 promotes metastasis through mutually regulating MUC5B expression in lung adenocarcinoma. Cell Death Dis. 2018;9(5):450. | ||
Ma T, Ma H, Zou Z, et al. The long intergenic noncoding RNA 00707 promotes lung adenocarcinoma cell proliferation and migration by regulating Cdc42. Cell Physiol Biochem. 2018;45(4):1566–1580. | ||
Zhang ZW, Chen JJ, Xia SH, et al. Long intergenic non-protein coding RNA 319 aggravates lung adenocarcinoma carcinogenesis by modulating miR-450b-5p/EZH2. Gene. 2018;650:60–67. | ||
Ding X, Zhang S, Li X, et al. Profiling expression of coding genes, long noncoding RNA, and circular RNA in lung adenocarcinoma by ribosomal RNA-depleted RNA sequencing. FEBS Open Bio. 2018;8(4):544–555. | ||
Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14(11):699–712. | ||
Han L, Zhang K, Shi Z, et al. LncRNA profile of glioblastoma reveals the potential role of IncRNAs in contributing to glioblastoma pathogenesis. Int J Oncol. 2012;40(6):2004–2012. | ||
Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum Mol Genet. 2010;19(R2):R152–R161. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



