Back to Journals » OncoTargets and Therapy » Volume 7
Expression profiles and initial confirmation of long noncoding RNAs in Chinese patients with pulmonary adenocarcinoma
Authors Zhao X, Zhu W, Zha W, Chen F, Wu Z, Liu Y, Huang M
Received 13 March 2014
Accepted for publication 2 May 2014
Published 1 July 2014 Volume 2014:7 Pages 1195—1204
DOI https://doi.org/10.2147/OTT.S64033
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Xin Zhao,* Wen Zhu,* Wangjian Zha, Feifei Chen, Zhenzhen Wu, Yanan Liu, Mao Huang
Department of Respiratory Medicine, The First Affiliated Hospital, Nanjing Medical University, Nanjing, People’s Republic of China
*These authors contributed equally to this study and share first authorship
Background: The purpose of this study was to investigate differentially expressed long noncoding RNAs (lncRNAs) in pulmonary adenocarcinoma tissue and adjacent noncancerous tissue from Chinese patients using lncRNA expression microarray and preliminary analysis.
Methods: RNA extracted from three paired pulmonary adenocarcinoma tissue and adjacent noncancerous tissue specimens was used to synthesize double-stranded complementary DNA after labeling and hybridization. The complementary DNA was labeled and hybridized to the lncRNA expression microarray, and array data were analyzed for hierarchical clustering. Gene coexpression networks were constructed to identify interactions among genes. To validate the microarray findings, we measured the relative expression levels of four random differentially expressed lncRNAs in the same tissue used for microarray using real-time quantitative polymerase chain reaction. The expression level of one lncRNA, AK124939, in the paired pulmonary adenocarcinoma/adjacent noncancerous tissue of another 30 patients was measured using real-time quantitative polymerase chain reaction. The experimental data were further analyzed and compared with clinical features.
Results: Of 39,000 lncRNAs investigated, 704 were differentially expressed in pulmonary adenocarcinoma tissue; 385 were upregulated and 319 were downregulated compared with those in the adjacent noncancerous tissue (fold change ≥2 and ≤–2, P<0.05). AK124939 expression levels in poorly differentiated adenocarcinoma tissue were lower than those found in well to moderately differentiated adenocarcinoma tissue (P=0.05).
Conclusion: There are significant differences in the lncRNA expression profiles in Chinese patients with pulmonary adenocarcinoma. LncRNAs such as AK124939 may be anticancer factors related to the progression of pulmonary adenocarcinoma.
Keywords: pulmonary adenocarcinoma, long noncoding RNA, microarray
Introduction
One of the greatest surprises in modern biology was the discovery that the human genome encodes only about 20,000 protein-coding genes, representing less than 2% of the total genome sequence.1 However, the advent of tiling resolution genomic microarrays and whole-genome and transcriptome sequencing technologies revealed that at least 90% of the genome is actively transcribed.2,3 It was found that the complexity of the human transcriptome exceeds that of a collection of protein-coding genes and their splice variants; there is extensive antisense, overlapping, and noncoding RNA expression.4–6
Although initially considered spurious transcriptional noise, recent evidence suggests that the so-called genomic “dark matter” may play a major biological role in cellular development and metabolism.7–10 The newly discovered long noncoding RNA (lncRNA) genes are one such element, with developmental and tissue-specific expression patterns and aberrant regulation in a variety of diseases, including cancer.11–19
Discovery of lncRNAs
LncRNAs were first described in a rat full-length complementary DNA library. LncRNAs were originally isolated in proximity to the noncoding RNA of protein-coding genes and were classified as sense, antisense, bidirectional, intronic, and intergenic according to their positions relative to the protein-coding genes in the genome. Initially, lncRNAs were considered transcriptions of genome “noise”, a byproduct of RNA polymerase II low-fidelity transcription with no biological function,9 and were thus neglected. In recent years, gradual recognition of the role of lncRNAs in X chromosome silencing, genomic imprinting, chromatin modification, transcription, transcription interference, and nuclear transport regulation have led to lncRNAs becoming a focus of research.
Biological characteristics and function of lncRNAs
LncRNAs are transcriptions of >200-nucleotide RNAs and are typically transcribed by RNA polymerase II. Besides temporal and spatial expression specificity, they lack meaningful open reading frames and do not code for proteins, instead directly regulating gene expression in the form of RNA at various levels, and are related to the occurrence and development of many diseases, including cancer.8
LncRNA genes are widely distributed in the genome and might be located in messenger RNA (mRNA) gene exons or introns, or between mRNA genes. At the same time, subcellular localization of lncRNAs is complex, as they can be located in the nucleus or cytoplasm.10 LncRNAs and mRNAs share many similar characteristics and overlapping areas: the genome “transcription hotspot” usually can be transcribed into both mRNA and lncRNA; each of the two chains of protein-coding genes can be transcribed into lncRNA;2 the same protein-coding gene can be transcribed into different mRNAs or lncRNAs; and a part of lncRNAs and mRNAs can be formed by alternative exon splicing combinations with polyadenylic acid in the 3′ terminal.
Generally, lncRNAs are mutable, but many contain highly conserved elements, which may be the result of a rapid adaptation to evolution20 that differs from that of mRNAs, which must ensure codon correctness to prevent open reading frame change. LncRNAs can only maintain a high level of conservation in sections that sustain secondary structure stability or those playing a key role in the functions of lncRNA. Studies have shown that mammalian lncRNAs are poorly conserved at the primary structure level while being highly conserved at secondary structure level.6,21
How lncRNAs regulate gene expression varies widely and is manifested in diverse lncRNA regulatory mechanisms. The effect of lncRNAs varies with the mechanism involved. LncRNAs can function as a main transcription factor, or a common regulatory that plays a role with other components. Gene expression regulation can generally be divided into:
- Regulation at the epigenetic modification level through interaction with various chromatin-modifying enzymes that modify chromatin by changing its conformation or activity, or repressing expression of the genes responsible. At the embryonic stage of development in particular, lncRNAs are involved in late silencing of allelic gene expression, maintaining epigenetic status, which is essential for normal development and cell differentiation in multicellular animals.
- Regulation at the transcriptional level through regulation of the combination and assembly of transcription factors, combination with regulatory DNA sequences to form three chain complexes, regulation of RNA polymerase II transcription, and transcription interference.
- Regulation at the post-transcriptional level through formation of double-stranded RNA with complementary mRNA, affecting mRNA processing, splicing, transportation, translation, and degradation.
Emerging role of lncRNAs in tumorigenesis
It has long been suspected that differential lncRNA expression plays a role in cancer; however, this suspicion has lacked strong supporting evidence. With advances in cancer transcriptome profiling and accumulating evidence supporting the function of lncRNAs, a number of differentially expressed lncRNAs have been associated with cancer. LncRNAs have been implicated in the regulation of a range of biological functions and the disruption of some of these functions, such as genomic imprinting and transcriptional regulation, thus playing a critical role in development of cancer.
Lung cancer is a malignant tumor with the highest morbidity and mortality worldwide. In 2003, Ji et al used sequencing and quantitative reverse transcription-polymerase chain reaction (qRT-PCR) and found that among 225 non-small cell lung cancer (NSCLC) patients, metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) lncRNA was overexpressed in 70 samples from patients with metastatic tumors. Further, the overexpression was stage-specific and tissue-specific, suggesting a close relationship between MALAT1 and the high metastatic potential and poor prognosis of NSCLC, thus implicating MALAT1 as a prognostic indicator of survival in patients with stage I NSCLC.22 The MALAT1 transcript is a >8,000-nucleotide noncoding RNA located on chromosome 11q13. Although it is expressed in normal human tissue and various tumors such as breast cancer, bladder cancer, liver cancer, and pancreatic cancer, overexpression in NSCLC is the most significant. Using cell migration experiments and qRT-PCR, Tano et al found that MALAT1-silencing mediated by small interfering RNA regulated the transcription and post-transcriptional expression of migration-related genes to affect cell migration ability in pulmonary adenocarcinoma, disrupting the migration of adenocarcinoma cells in vitro.23 These studies indicate that MALAT1 regulates metastasis. In addition, using overexpression and small interfering RNA interference methods, Tripathi et al found that MALAT1 and serine/arginine-rich (SR) proteins interact to affect the distribution of SR and other splicing factors in small nuclear areas. MALAT1 affected alternative splicing of the pre-mRNA precursor by regulating the level of phosphorylated SR protein,24 which is of great significance in the study of SR protein splicing regulation, although its role in tumorigenesis remains unknown.
An important lung cancer, pulmonary adenocarcinoma is one of the most common human cancers, with poor 5-year survival rates worldwide. In this study, we profiled lncRNA expression in Chinese patients with pulmonary adenocarcinoma and performed a preliminary analysis of the experimental and clinical data.
Materials and methods
Samples
Three paired pulmonary adenocarcinoma tissue (T1, T2, T5) and adjacent noncancerous tissue (N1, N2, N5) specimens from patients with grade II tumors and without a history of chemotherapy and radiotherapy were used for the microarray study. The tumor (T) and normal (N) tissues were stored in liquid nitrogen. The fourth T/N tissue pair used in the microarray was a mixture of the first three pairs: T6 was a combination of T1, T2, and T5, and N6 was a combination of N1, N2, and N5.
Another 30 paired pulmonary adenocarcinoma/adjacent noncancerous tissue specimens from patients who had undergone surgical therapy at the First Affiliated Hospital of Nanjing Medical University between July 2012 and February 2013 and for which the postoperative pathological diagnosis was confirmed to be pulmonary adenocarcinoma were obtained for the qRT-PCR study.25,26 All specimens were collected after patients had signed informed consent forms and following review by the hospital ethics committee.
Microarray and computational analysis
RNA extracted from the three paired tissue specimens was used to synthesize double-stranded complementary DNA after labeling and hybridization. The complementary DNA was labeled and hybridized to an Agilent Human lncRNA + mRNA array version 2.0 (Agilent Technologies, Santa Barbara, CA, USA) designed with four identical arrays per slide (4 × 180,000 format). Each array contained probes interrogating 39,000 human lncRNAs and 32,000 human mRNAs. Each RNA was detected twice by the probes. The array also contained 4,974 Agilent control probes.
The array data were analyzed for summarization, normalization, and quality control using GeneSpring software version 11.5 (Agilent). To select differentially expressed genes, we used threshold values of a ≥2- and ≤−2-fold change and a Benjamini–Hochberg corrected P-value of 0.05. The data were log2 transformed and median-centered by genes using the Adjust Data function of CLUSTER version 3.0 software (Michiel de Hoon, Human Genome Center, University of Tokyo, Tokyo, Japan), then further analyzed with hierarchical clustering with average linkage.27 Lastly, we performed tree visualization using Java TreeView (Stanford University School of Medicine, Stanford, CA, USA).
The microarray data discussed in this paper have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus and can be accessed through the series accession number GSE56850 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE56850).
Coexpression network
Gene coexpression networks are used to identify interactions among genes28 according to the normalized signal intensity of specific expressed genes. We constructed network adjacency between two genes, i and j, defined as a power of the Pearson correlation between the corresponding gene expression profiles xi and xj. We then obtained a gene adjacency matrix, M (i,j),29 by computing the profiles, visualized as a graph, the topological properties of which were examined. To construct a visual representation, only the strongest correlations (≥0.99) were drawn in the renderings. Each gene corresponded to a node in the gene coexpression networks. Two genes connected by a line indicated a strong correlation (ie, red lines denote positive correlation, blue lines denote negative correlation.). In network analysis, a degree is the simplest, most important measure of the centrality of a gene within a network and determines the relative importance. A degree is defined as the number of directly linked neighbors.30
qRT-PCR
The qRT-PCR was performed using PrimeScript RT Master Mix (Takara, Otsu, Japan) and SYBR Green Mix (ABI, Foster City, CA, USA). Total RNA was extracted from tissues using TRIzol (Invitrogen, Carlsbad, CA, USA) and purified with a mirVana miRNA isolation kit (Ambion, Austin, TX, USA) according to the manufacturer’s protocol. RNA purity and concentration were determined from optical density OD260/280 readings using a spectrophotometer (NanoDrop ND-1000; Thermo Fisher Scientific, Wilmington, DE, USA); RNA integrity was determined by 1% formaldehyde denaturing gel electrophoresis. The RNA was reverse-transcribed into complementary DNA by combining 4 μL 5× PrimeScript RT Master Mix, 2 μL template RNA, and RNase-free distilled water to a total volume of 20 μL in a microtube, heating at 37°C for 15 minutes, then at 85°C for 5 seconds, followed by immediate cooling on ice. The purity and concentration of the complementary DNA were determined from OD260/280 readings using the NanoDrop ND-1000 spectrophotometer. Each 10 μL reaction contained 5 μL 2× SYBR Select Master Mix, 0.2 μL each forward and reverse primers, a complementary DNA template (up to 100 ng of complementary DNA was used in each reaction for optimal performance), and 4.6 μL of RNase-free distilled water, and was transferred to wells in an optical plate. The plate was sealed with an optical adhesive cover and centrifuged briefly to spin down the contents and eliminate air bubbles before being placed in a StepOne thermocycler (Life Technologies, Carlsbad, CA, USA). The thermal cycling conditions were as follows: uracil-DNA glycosylase activation at 50°C for 2 minutes, AmpliTaq DNA polymerase (Life Technologies) and primer activation at 95°C for 2 minutes, denaturation at 95°C for 15 seconds, and annealing/extension at 60°C for one minute for 40 cycles. We programmed the instrument to perform a default dissociation step. We used glyceraldehyde-3-phosphate dehydrogenase as the internal control throughout the study.
Statistical analysis
All statistical analyses were performed using Statistical Package for the Social Sciences version 20 software (SPSS Inc., Chicago, IL, USA). We used one-way analysis of variance, Fisher’s exact test, Pearson correlation analysis, and the two-tailed t-test (including the independent-samples t-test and paired t-test). Statistical significance was set at a P-value of <0.05.
Results
LncRNA expression profiles in pulmonary adenocarcinoma
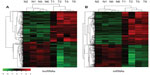
Of 39,000 lncRNAs, 704 were differentially expressed in pulmonary adenocarcinoma tissue; 385 were upregulated and 319 were downregulated as compared with the adjacent noncancerous tissue (fold change ≥2 and ≤−2, P<0.05). Hierarchical clustering revealed systematic variations between the expression of lncRNAs and protein-coding RNAs in the paired pulmonary adenocarcinoma/nontumor lung tissue specimens (Figure 1A and B). The data indicate that a series of lncRNAs is frequently differentially expressed in pulmonary adenocarcinoma tissues.
Bioinformatics analysis and validation of lncRNA expression profiles
Gene coexpression networks are used to cluster thousands of transcripts into phenotypically relevant coexpression modules.28,31 Given that coexpression networks may correspond to biological pathways32 and many functions of protein-coding RNAs are present in the National Center for Biotechnology Information Reference Sequence database, we concentrated on coexpression with a high rate of protein-coding RNAs in the carcinoma coexpression network, selecting four lncRNAs (Table 1), ie, AK124939, ASO3690, ENST00000491282, and XLOC_000575, in pulmonary adenocarcinoma for characterization using this method. In the cancer coexpression network, AK124939 was connected to a protein-coding gene, PDZD2 (also known as AIPC, PIN1, PAPIN, PDZK3), which is involved in tumorigenesis and tumor development. ASO3690 and XLOC_000575 were each connected to two protein-coding genes; ENST00000491282 was connected to 17 protein-coding genes (Figure 2).
  | Table 1 The four studied long noncoding RNAs |
To validate the microarray findings, we analyzed the expression of the abovementioned lncRNAs with qRT-PCR in the same three paired pulmonary adenocarcinoma tissue/adjacent noncancerous tissue specimens used for the microarray. Table 2 lists the corresponding primer sequences. The data confirmed that ASO3690, ENST00000491282, and XLOC_000575 were overexpressed in pulmonary adenocarcinoma, whereas AK124939 expression was decreased (P<0.05 for all, Figure 3). These results were sufficiently consistent with that of the lncRNA expression profiles.
  | Table 2 Primer sequences |
AK124939 may be related to progress of pulmonary adenocarcinoma
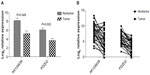
To explore the correlation between lncRNA and pulmonary adenocarcinoma, we increased the number of samples studied by obtaining 30 paired pulmonary adenocarcinoma/adjacent noncancerous tissue specimens and detected AK124939 and its related PDZD2 mRNA (Table 2 lists the primer sequences) using qRT-PCR. The transcript levels of AK124939 and PDZD2 mRNA were both lower in pulmonary adenocarcinoma tissues (P<0.05) compared with the paired noncancerous lung tissues (Figure 4A and B). Pearson correlation analysis estimated r=0.741 (P<0.05) in the tumor tissues and r=0.905 (P<0.05) in the adjacent noncancerous tissues (Figure 5A and B). Therefore, there was a clear positive correlation between the lncRNA AK124939 and PDZD2 mRNA.
We then explored the relationship between level of AK124939 expression and the clinical and pathological characteristics of patients with pulmonary adenocarcinoma. There was no significant correlation between AK124939 expression and age, sex, tumor-node-metastasis (TNM) stage, tumor size, Karnofsky performance status, smoking history, or chemotherapy history. Although not statistically significant (P=0.05), the level of AK124939 expression was related to tumor differentiation grade. AK124939 expression in the poorly differentiated adenocarcinoma tissue was lower than that in well to moderately differentiated adenocarcinoma tissue (Table 3).
  | Table 3 Patient characteristics |
Discussion
Once considered transcription “noise”, the possible role of lncRNAs in cancer development and occurrence has recently been revealed, for which studies are ongoing. Previous studies reported more significant MALAT1 lncRNA overexpression in NSCLC compared with that in other cancers22 and that lncRNA is a possible regulatory factor of metastasis.23 Given these previous findings, we aimed to determine whether the same would hold true for other lncRNAs in pulmonary adenocarcinoma, which has been described as the most prevalent form of NSCLC.33
We used a commercial microarray and qRT-PCR to investigate the differential expression of lncRNAs in pulmonary adenocarcinoma tissue specimens and compared them with that in adjacent noncancerous tissue specimens. Analysis of the microarray data revealed significant differential expression for 704 lncRNAs (385 upregulated and 319 downregulated) in pulmonary adenocarcinoma tissue compared with that in adjacent normal tissue. Subsequent gene coexpression network analysis revealed that the coexpression patterns of the lncRNAs and protein-coding RNAs of the pulmonary adenocarcinoma tissue differed from that in normal tissue.
We characterized the lncRNAs, AK124939, ASO3690, ENST00000491282, and XLOC_000575, which had a high rate of protein-coding RNAs in the carcinoma coexpression network, and found that all but AK124939 were upregulated in pulmonary adenocarcinoma tissue. Similarly, mRNA expression of the tumor suppressor gene PDZD2, related to AK124939, was downregulated, indicating a correlation between the two. It has been demonstrated that PDZD2 is upregulated in primary prostate tumors and may be involved in the early stages of prostate tumorigenesis.34,35 In light of these findings, more research is warranted to explore the difference in the mechanisms and relationship between lncRNAs and protein-coding RNAs.
Every pattern of pulmonary adenocarcinoma is assigned a grade related to its metastatic or recurrence potential: grade 1, lepidic; grade 2, acinar or papillary; and grade 3, solid or micropapillary, which correspond to low, intermediate, and high metastatic potential, respectively. Well differentiated tumors (histologic score 3, lepidic pattern) are associated with low rates of recurrence, while poorly differentiated tumors (histologic score of 5 and 6, solid or micropapillary pattern) are associated with higher rates of recurrence. Moderately differentiated tumors with a histologic score of 4 have pure acinar, papillary, or combined acinar and papillary as the predominant and secondary predominant patterns.36,37 Among the 30 cases, four (13.3%) were well differentiated (grade 1/G1), ten (33.3%) were moderately differentiated (grade 2/G2), and 16 (53.3%) were poorly differentiated (grade 3/G3).
Further investigation of AK124939 expression in the 30 paired pulmonary adenocarcinoma/adjacent noncancerous tissues revealed that its expression levels in the poorly differentiated adenocarcinoma tissue were lower than that in well to moderately differentiated adenocarcinoma tissue (albeit P=0.05). Given the low number of cases and lack of proportion, we merged G1 and G2 into a well to moderately differentiated group and compared these with the poorly differentiated (G3) group; there were no significant differences between G1 or G2 and G3. It is known that the poorer the tumor cell differentiation, the higher the degree of malignancy. In patients with the same TNM stage, histopathological grade tends to have an effect on prognosis. It has been shown that lncRNAs are significant regulators of gene expression that interact with the major pathways of cell growth, proliferation, differentiation, and survival.38,39 Alterations in the function of lncRNAs promote tumor formation and progression, and metastasis; for example, noncoding Nras functional RNA is involved in malignant transformation of colonic epithelial cells.40 Therefore, if future studies involving a larger sample size confirm that there is a significant difference (P<0.05), we may assume that AK124939 is related to cell differentiation and tumor formation as a suppressor gene. From this perspective, multicenter, randomized concurrent controlled studies are required to determine the potential of AK124939 as a diagnostic and prognostic marker. Our findings indicate that, like MALAT1 lncRNA, AK124939 plays a potential role in targeted treatment of lung cancer, or as a marker of diagnosis or prognosis, and further research is required.
Currently, a series of issues remain to be clarified. For example, it is suggested that a large number of lncRNAs detected in this study, ie, those other than AK124939, ASO3690, ENST00000491282, and XLOC_000575, are aberrantly expressed in pulmonary adenocarcinoma tissues. Thus, it remains to be determined whether these lncRNAs are strongly correlated with survival in pulmonary adenocarcinoma and if they can be used as biomarkers for early clinical diagnosis.
Clearly, there is much work to be done. RNA pull-down assays are indispensable both in vitro and in vivo. Moreover, describing the molecular mechanisms by which lncRNAs function in pulmonary adenocarcinoma is critical for elucidation of their role in cancer occurrence and development. Thus, lncRNAs have potential use as diagnostic and prognostic markers in pulmonary adenocarcinoma and in the development of lncRNA-mediated therapy.
Conclusion
We detected lncRNAs that are differentially expressed in pulmonary adenocarcinoma tissue compared with paired adjacent noncancerous tissue from Chinese patients. AK124939 was downregulated in pulmonary adenocarcinoma compared with nontumor tissues. Moreover, our results indicate that AK124939 levels in poorly differentiated adenocarcinoma tissue are lower than those in well to moderately differentiated adenocarcinoma tissue. In addition, gene coexpression networks revealed a positive correlation between PDZD2 mRNA and AK124939. Taken together, these lncRNA expression profiles in Chinese patients with pulmonary adenocarcinoma indicate that lncRNAs have potential use as diagnostic and prognostic markers in pulmonary adenocarcinoma and in the development of lncRNA-mediated therapy.
Acknowledgment
This work was supported by a grant from the Jiangsu Provincal Special Program of Medical Science (BL2012012).
Disclosure
The authors report no conflicts of interest in this work.
References
Ponting CP, Belgard TG. Transcribed dark matter: meaning or myth? Hum Mol Genet. 2010;19(R2):R162–R168. | |
Birney E, Stamatoyannopoulos JA, Dutta A, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. | |
Costa FF. Non-coding RNAs: meet thy masters. Bioessays. 2010;32(7):599–608. | |
Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8(6):413–423. | |
Khachane AN, Harrison PM. Mining mammalian transcript data for functional long non-coding RNAs. PLoS One. 2010;5(4):e10316. | |
Guttman M, Amit I, Garber M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–227. | |
van Bakel H, Nislow C, Blencowe BJ, Hughes TR. Most “dark matter” transcripts are associated with known genes. PLoS Biol. 2010;8(5):e1000371. | |
Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–1504. | |
Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–641. | |
Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. | |
Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. | |
Loewer S, Cabili MN, Guttman M, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42(12):1113–1117. | |
Castle JC, Armour CD, Lower M, et al. Digital genome-wide ncRNA expression, including SnoRNAs, across 11 human tissues using polyA neutral amplification. PLoS One. 2010;5(7):e11779. | |
Wu SC, Kallin EM, Zhang Y. Role of H3K27 methylation in the regulation of lncRNA expression. Cell Res. 2010;20(10):1109–1116. | |
Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J Pathol. 2010;220(2):126–139. | |
Maruyama R, Shipitsin M, Choudhury S, et al. Breast cancer special feature: altered antisense-to-sense transcript ratios in breast cancer. Proc Natl Acad Sci U S A. 2012;109(8):2820–2824. | |
Silva JM, Perez DS, Pritchett JR, Halling ML, Tang H, Smith DI. Identification of long stress-induced non-coding transcripts that have altered expression in cancer. Genomics. 2010;95(6):355–362. | |
Perez DS, Hoage TR, Pritchett JR, et al. Long, abundantly expressed noncoding transcripts are altered in cancer. Hum Mol Genet. 2008;17(5):642–655. | |
Guffanti A, Iacono M, Pelucchi P, et al. A transcriptional sketch of a primary human breast cancer by 454 deep sequencing. BMC Genomics. 2009;10:163. | |
Pang KC, Frith MC, Mattick JS. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 2006;22(1):1–5. | |
Washietl S, Hofacker IL, Lukasser M, Huttenhofer A, Stadler PF. Mapping of conserved RNA secondary structures predicts thousands of functional noncoding RNAs in the human genome. Nat Biotechnol. 2005;23(11):1383–1390. | |
Ji P, Diederichs S, Wang W, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22(39):8031–8041. | |
Tano K, Mizuno R, Okada T, et al. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010;584(22):4575–4580. | |
Tripathi V, Ellis JD, Shen Z, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39(6):925–938. | |
Ceelen L, De Craene J, De Spiegelaere W. Evaluation of normalization strategies used in real-time quantitative PCR experiments in HepaRG cell line studies. Clin Chem. 2014;60(3):451–454. | |
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. | |
Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95(25):14863–14868. | |
Pujana MA, Han JD, Starita LM, et al. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat Genet. 2007;39(11):1338–1349. | |
Prieto C, Risueno A, Fontanillo C, De las Rivas J. Human gene coexpression landscape: confident network derived from tissue transcriptomic profiles. PLoS One. 2008;3(12):e3911. | |
Barabasi AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5(2):101–113. | |
Liao Q, Liu C, Yuan X, et al. Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co-expression network. Nucleic Acids Res. 2011;39(9):3864–3878. | |
Nayak RR, Kearns M, Spielman RS, Cheung VG. Coexpression network based on natural variation in human gene expression reveals gene interactions and functions. Genome Res. 2009;19(11):1953–1962. | |
Hoenerhoff MJ, Hong HH, Ton TV, Lahousse SA, Sills RC. A review of the molecular mechanisms of chemically induced neoplasia in rat and mouse models in National Toxicology Program Bioassays and their relevance to human cancer. Toxicol Pathol. 2009;37(7):835–848. | |
Chaib H, Rubin MA, Mucci NR, et al. Activated in prostate cancer: a PDZ domain-containing protein highly expressed in human primary prostate tumors. Cancer Res. 2001;61(6):2390–2394. | |
Tam CW, Liu VW, Leung WY, Yao KM, Shiu SY. The autocrine human secreted PDZ domain-containing protein 2 (sPDZD2) induces senescence or quiescence of prostate, breast and liver cancer cells via transcriptional activation of p53. Cancer Lett. 2008;271(1):64–80. | |
Sica G, Yoshizawa A, Sima CS, et al. A grading system of lung adenocarcinomas based on histologic pattern is predictive of disease recurrence in stage I tumors. Am J Surg Pathol. 2010;34(8):1155–1162. | |
Moreira AL, Joubert P, Downey RJ, Rekhtman N. Cribriform and fused glands are patterns of high-grade pulmonary adenocarcinoma. Hum Pathol. 2014;45(2):213–220. | |
Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. 2012;9(6):703–719. | |
Hiriart E, Verdel A. Long noncoding RNA-based chromatin control of germ cell differentiation: a yeast perspective. Chromosome Res. 2013;21(6–7):653–663. | |
Franklin JL, Rankin CR, Levy S, et al. Malignant transformation of colonic epithelial cells by a colon-derived long noncoding RNA. Biochem Biophys Res Commun. 2013;440(1):99–104. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.