Back to Journals » OncoTargets and Therapy » Volume 11
Expression of protein disulfide isomerase A3 precursor in colorectal cancer
Authors Yang Z, Liu J, Shi Q, Chao Y, Di Y, Sun J, Zhang J, Huang L, Guo H, He C
Received 18 October 2017
Accepted for publication 17 April 2018
Published 19 July 2018 Volume 2018:11 Pages 4159—4166
DOI https://doi.org/10.2147/OTT.S154452
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Zhaowei Yang,1 Juan Liu,2 Qinghong Shi,1 Yang Chao,3 Yanqing Di,3 Jing Sun,3 Jiebing Zhang,3 Lihong Huang,4 Honghua Guo,3 Chengyan He1
1Laboratory Medicine Center, China-Japan Union Hospital of Jilin University, Changchun, China; 2Department of Digestive Internal Medicine, Qian Wei Hospital of Jilin Province, Changchun, China; 3Department of Digestive Internal Medicine, China-Japan Union Hospital of Jilin University, Changchun, China; 4Department of Geriatrics, China-Japan Union Hospital of Jilin University, Changchun, China
Introduction: Over 20% of colorectal cancer (CRC) patients seek medical attention for the first time when they are in the advanced stages of CRC. Thus, early and reliable detection of CRC is critical to early diagnosis of CRC. Protein disulfide isomerase A3 precursor (PDIA3) has been implicated in various types of cancers. However, little is known about PDIA3 in CRC.
Methods: In this study, we screened PDIA3 expression in CRC tissues and cell lines. Small interfering RNA (siRNA) was introduced into SW480 cells to knockdown PDIA3 expression. The effect of PDIA3 in cell growth was evaluated.
Results: Significant upregulation of PDIA3 expression was found in CRC tissues as compared with adjacent non-cancer tissues, and was found in established CRC cell lines (SW480, HCT116, CACO2, NCM460 and HT-29). In SW480 cells, knockdown of PDIA3 expression with siRNA resulted in subcellular morphological change, reduced cell proliferation and increased apoptosis.
Conclusion: PDIA3 inhibition could suppress CRC, likely through inducing apoptosis. PDIA3 could be a potential therapeutic target for CRC.
Keywords: PDIA3, colorectal cancer, proteomics, siRNA, apoptosis
Introduction
Colorectal cancer (CRC) is the third most prevalent malignancy in developed countries.1 The incidence of CRC is about 13.29 per 100,000 people in China and has been rising year by year.2 Although great progress has been achieved in the diagnosis and therapy of cancer, CRC is still the second most common cause of cancer-related death in men and the third most common cause in women.3 Tumor resection is considered to be the optimal choice, and the 5-year survival rate is up to 90% in CRC patients without lymph node metastasis.4 Early detection is critical for favorable prognosis in cancer treatment. Therefore, molecular biomarker profiling and validation are the key issues for early cancer diagnosis.5
Protein disulfide isomerase A3 precursor (PDIA3), also known as Erp57 and Erp60, is a member of the protein disulfide isomerase (PDI)-like family.6 It is a 58 kDa multifunctional protein and is located mostly in the endoplasmic reticulum (ER) lumen where it functions as a component of glycoprotein-specific quality control machinery to participate in the folding of numerous cysteine-rich glycoproteins.6 PDIA3 also plays important roles in cell signal transduction, major histocompatibility complex (MHC) assembly,7,8 platelet function,9 transcription,10 sperm egg fusion,11 bone development12 and apoptosis induction. In addition, PDIA3 was implicated in oncogenesis and tumor progression.13,14 However, the role of PDIA3 in cancer occurrence and progression is controversial. One study reported that downregulated PDIA3 expression is associated with poor prognosis of early stage cervical cancer.15 Meanwhile, another study revealed that PDIA3 upregulation was involved in breast carcinogenesis.16 PDIA3 is considered as a component of the signal transducer and activator of transcription 3-transcriptional complex.17 Downregulation of PDIA3 disrupted the Wnt signaling pathway by reducing beta-catenin stability in cervical cancer cells.18 Apparently, the aforementioned signaling pathways are critical in tumorigenesis, cell proliferation and apoptosis, tumor metastasis, angiogenesis and chemoresistance.19,20 However, the relationship between PDIA3 expression and CRC remains largely unknown. In this study, we initially investigated the expression of PDIA3 in CRC tissues and cell lines and studied the effect of PDIA3 inhibition on CRC cells.
Materials and methods
Tissue specimens
CRC and adjacent non-cancerous tissue specimens (about 10 cm away from tumor edge) were obtained from 23 treatment-naïve patients with pathologically proven TNM-III colorectal adenocarcinoma who underwent tumor resection at the Department of Oncology, China-Japan Union Hospital of Jilin University (Changchun, China) between February 2015 and February 2016. The tissue specimens were snap frozen and stored in liquid nitrogen. Written informed consent was obtained from the patients. The study protocol was approved by the ethics committee of China-Japan Union Hospital of Jilin University.
Protein extraction
The frozen tissues were cut into small pieces and homogenized in liquid nitrogen. The homogenized samples were then extracted with ice-cold ReadyPrep protein extraction kit (Bio-Rad, Hercules, CA, USA) for 20 minutes. The supernatant was collected after centrifugation at 12,000× g for 4 minutes. The protein concentrations were determined using a bicinchoninic acid protein assay kit (Abcam, Rockford, IL, USA). The supernatants were stored at −80°C before use.
Proteolysis
The supernatants were thawed at room temperature and dissolved in 3–5 volumes (v/v) NH4HCO3 (50 mM) solution. For protein solubilization and denaturation, the samples were incubated with 20 mM DL-Dithiothreitol-containing NH4HCO3 at 56°C for 1 hour, and then with 50 mM iodoacetamide-containing NH4HCO3 at room temperature for 30 minutes. The samples were then incubated with gold grade trypsin (Promega, Madison, WI, USA) overnight at 37°C.
Mass spectrometry and two-dimensional hyphenated chromatographic analysis
The samples were dissolved in buffer A (0.1% formaldehyde). A 50 mg sample was separated in a strong cation exchange column (150 × 0.32 mm) and reversed phase column (150 × 0.17 mm). An strong cation-exchange capillary column was applied in ammonium acetate strong cation column elution stage following a gradient of 0 mM, 25 mM, 50 mM, 75 mM, 100 mM, 125 mM, 150 mM and 1 M. The eluted peptides were then transferred onto a reverse phase capillary column (PepMap C18, 75 μm ID ×150 mm, 3 μm particle and 100 Å pore size; Dionex, Amsterdam, the Netherlands). The elution gradient for the reversed phase column was 30% buffer B (0.1% formic acid, 99.9% acetonitrile) over 3 hours at a flow rate of 2 μL/min. A general scan by Mass range m/z 400–2000 was followed by 10 data-dependent MS/MS scans (10 μ scans each, isolation width 3 amu, 35% normalized collision energy, dynamic exclusion for 1.5 minutes) on an electrospray linear ion trap mass spectorphotemeter (LTQ XL) (Thermo Fisher Scientifc, San Jose, CA, USA).
The raw files containing MS/MS spectra were searched against the SwissProt protein database using SEQUEST algorithm that was built in BioWorks 3.3.1 SP1 (Thermo Fisher Scientific). The results were filtered to obtain positive identifications by the following criteria: Rsp was 1; Delta Cn was at least 0.19; X corr was 2.2 for single charged peptides, 2.5 for doubly charged peptides, 2.9 for triply charged peptides. In this study, protein abundance was evaluated based on the number of MS/MS spectra of a certain protein in different samples. When the number of MS/MS spectra of a certain protein identified in CRC tissues was statistically larger than that of the same protein in adjacent non-cancer tissues, this protein was considered to be upregulated.
Cell culture
SW480, HCT116, CACO2, NCM460 and HT-29 cell lines were purchased from the Cell Bank of Chinese Academy of Sciences. Cells were cultured in complete medium (Dulbecco’s Modified Eagle’s Medium) with 5% CO2 at 37°C. Cells were seeded on the round cell slide until the cells grew to 60%–90% confluence and were washed with Hank’s solution.
Construction of PDIA3 siRNA vector
SW480 and HCT116 cells were inoculated in 6-well plates and were transfected when they were 90%–95% confluent. Two small interfering RNAs (siRNAs; Santa Cruz Biotechnology, Santa Cruz, CA, USA) were designed to knockdown PDIA3 expression: siRNA-1: 5’-GGACAAGACUGUGGCAUAUTTAUAUGCCACAGUCUUGUCC TT-3’; siRNA-2: 5’-CAGCCAACAAGAAGCUAAATTUUUAGCUUCUUGUUG GCUGTT-3’. PDIA3 siRNA vector (4 μg) was diluted in the 250 mL Opti-MEM low serum medium and mixed gently. Lipofectamine TM 2000 (Huiji Biotechnology Co., Hangzhou, China) was used to assist transfection. Cells were cultured at 37°C with 5% CO2 for 6 hours, and then the medium was replenished over a period of 48 hours. Transfection efficiency was confirmed by immunofluorescence staining and western blotting assays.
Western blotting assays
PDIA3 expression in CRC and adjacent non-cancerous tissues was also confirmed by western blotting assays. SW480 and HCT116 cells were incubated with siRNAs for 24 or 48 hours. Thereafter, the cells were collected, washed with PBS and lysed with radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime, Shanghai, China). The cell lysates were clarified by centrifugation at 12,000 rpm for 15 minutes. Protein samples were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis. After electrophoresis, the proteins were transferred onto nitrocellulose membranes. After blocking in 5% de-fat milk, the membranes were probed with primary antibodies against PDIA3 (1:400; Sangon, Shanghai, China) and β-actin (1:1,000; Sangon). Protein bands were detected using fluorescence conjugated secondary antibody (1:2,000, A0216; Beyotime) and visualized with the chemiluminescence method.
Immunofluorescence staining
When SW480 cells were 80% confluent in the glass-covered slips, they were washed with Hank’s solution 3 times, and fixed with 4% paraformaldehyde for 30 minutes at room temperature. The cells were washed 3 times with PBS, and incubated with 0.1% Triton X-100 at room temperature for 10 minutes. The slides were blocked with 5% goat serum at room temperature for 1 hour, and incubated with rabbit polyclonal anti-PDIA3 antibody (SAB, 1:100) for 1 hour, followed by incubation with Alexa555-labeled goat anti-rabbit secondary antibodies (Molecular Probe, 1:200) for 1 hour at room temperature. The nuclei were stained by Hoechest33342 for 5 minutes. PDIA3 expression was observed under a fluorescence microscope and imaged by confocal microscopy.
Quantitative real-time polymerase chain reaction (PCR)
SW480 cells were incubated with siRNAs for 24 or 48 hours. Thereafter, the cells were collected and total RNA was isolated using Trizol Reagent (Invitrogen, Carlsbad, CA, USA) and reversely transcribed into cDNA using all-in-one first-strand cDNA synthesis kit (Genecopoeia, Guangzhou, China) according to the manufacturers’ protocols. PDIA3 expression was quantified by real-time PCR method using the real-time PCR master mix kit (Genecopoeia). β-Actin was used as an internal reference. The sequences of the primers used were as follows: β-actin-forward: 5′-CATGTACGTTGCTATCCAGGC-3′, β-actin-reverse: 5′-CTCCTTAATGTCACGCACGAT-3′; PDIA3-forward: 5′-GCCTCCGACGTGCTAGAAC-3TGPDIA-3′, PDIA3-reverse: 5′-GCGAAGAACTCGACGAGCAT-3′.
CCK-8 assays
Cell proliferation was assayed by using Cell Counting Kit-8 (CCK-8) (Dojindo, Kumamoto, Japan). SW480 cells were digested with 0.25% trypsin before transfection, and then inoculated in 48-well plates. PDIA3-siRNA vector (0–0.8 μg) was transfected in triplicates. Cells were cultured at 37°C with 5% CO2 for 48 hour. In total, 50 μL CCK-8 solution was added into each well. Absorbance (A) was read at 450 nm wavelength using a microplate reader. Inhibition rate = (1 − AExperimental group/AControl) × 100%. Cellular morphology was imaged under an optical microscope.
Electron microscopy
SW480 cells were fixed in 1% osmium tetroxide in 0.1 M phosphate buffer for 1 hour. Images were taken under the electron microscope (Hitachi, Tokyo, Japan).
Apoptosis assays
SW480 cells were cultured in 4–6 cm culture dishes and transfected with PDIA3 siRNA vector and then cultured at 37°C with 5% CO2 for 48 hours. SW480 cells were digested by 0.25% trypsin, and then detected with Annexin V-PE cell apoptosis detection kit (Beyotime) in accordance with the instruction. The specimens were cover-slipped with anti-fade solution and observed under the fluorescence microscope. The excitation wavelength range was 450–500 nm, and the emission wavelength range was 515–565 nm (green fluorescence).
Statistical analysis
Data were analyzed with Student’s t-test using SPSS software, version 13.0 (SPSS Inc., Chicago, IL, USA). P-values less than 0.05 were considered statistically significant.
Results
Total protein profiles in CRC and adjacent non-cancer tissues
The protein expression profile of CRC and adjacent non-cancer tissues was determined using two-dimensional chromatography and MS. The results showed that 15 types of proteins (including PDIA3) were upregulated and 9 proteins were downregulated in CRC tissues compared with adjacent non-cancer tissues. The upregulated proteins were MYH9, TPM4, HIST1H3J, NPM1, FGA, GSTP1, IGHA1, TPM3, IGHA2, PDIA3, KRT18, TUBB, PKM2, IGHV3OR16-13 and HIST1H2AE, and the downregulated proteins were SYNM, MYH11, VCL, FLNC, TUBB2A, MYL6, TLN1, ACTC1 and CNN1. MS revealed the results of PDIA3 (Figure 1).
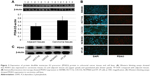
Expression of PDIA3 protein in tumor tissues and cell lines
Western blotting analysis showed a significant increase of PDIA3 expression in CRC compared with adjacent non-cancer tissues (Figure 2A). PDIA3 expression in several CRC cell lines, including SW480, HCT116, CACO2, NCM460 and HT-29, was determined using western blotting and immunofluorescence assays. A robust cytoplasmic expression of PDIA3 was observed in all cell lines (Figure 2B). Importantly, SW480 cells had the highest expression of PDIA3 (Figure 2B and C).
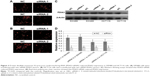
In the subsequent experiments, we investigated the role of PDIA3 in the growth of CRC cells. To further establish a correlation between PDIA3 expression and cell viability, we performed small interference RNA assays. SW480 and HCT116 cells were transfected with siRNA-PDIA3 vector, and PDIA3 expression was measured by immunofluorescence and western blotting assays. We found that both siRNA-PDIA3 vectors significantly reduced PDIA3 expression in SW480 and HCT116 cells (Figure 3).
Knockdown of PDIA3 expression reduced the proliferation of SW480 cells
Effect of PDIA3 expression on SW480 cell proliferation was confirmed by CCK-8 assay. Incubation with both siRNA-PDIA3 vectors inhibited cell viability dose and time dependently (Figure 4A and B).
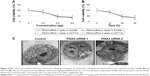
Furthermore, the effect of siRNA-PDIA3 on cell morphological changes was detected by transmission electron microscopy. As shown in Figure 4C, incubation with siRNA-PDIA3 obviously changed cellular morphology, membrane thickness and mitochondria shapes in SW480 cells.
Knocking down PDIA3 expression induced apoptosis of SW480 cells
SW480 cells were treated with siRNA-PDIA3 for 48 hours. The proportion of apoptotic cells (green) was significantly increased after PDIA3 knockdown (27.8% ± 8.59%) (Figure 5A) compared with the control group (9.8% ± 2.58%) (P<0.01) (Figure 5B).
We also detected Bcl-2 and Bax expression. As shown in Figure 5C–E, siRNA-PDIA3 significantly promoted Bax expression, while reducing Bcl-2 expression.
Discussion
Early accurate diagnosis of CRC is closely associated with the prognosis of CRC patients.15 Therefore, selecting molecular biomarkers that are closely related to the occurrence and progression of CRC has been pursued. In this study, we found that PDIA3 expression in CRC tissues and several CRC cell lines was unregulated, while siRNA-mediated knockdown of PDIA3 resulted in the death of CRC cells via an apoptotic pathway.
PDIA3 is a member of the PDI family that acts as chaperones and is localized in the ER, cytosol and nucleus.14 It was reported that PDIA3 was significantly upregulated in uterine cancer, lung cancer and gastric cancer,21 and even PDIA3 was proposed to be a potential tumor marker for uterine cancer.22 The oncogenic function of PDIA3 can be explained by its interaction with relevant signaling pathways. For example, PDIA3 directly binds with DNA fragments or functions as an integral component of the MHC class I pathway to influence gene expression.7 PDIA3 was also reported to be downregulated in certain type of cancers, especially breast cancer.16 Additionally, PDIA3 expression was found to be associated with drug resistance in epithelial ovarian cancer.13 It is also found that the regulation of PDIA3 for transformation/transcription domain-associated protein (TRRAP) is reduced following resveratrol treatment.10 TRRAP, a potential cofactor for C-myc,23 functions as a scaffold for the assembly of proteins in the process of cell transformation.
In this study, we demonstrated for the first time the upregulation of PDIA3 levels in CRC tissues. Our results suggested that PDIA3 might be a candidate CRC biomarker. As PDIA3 knockdown could induce apoptosis of ovarian and breast cancer cells,16 we asked whether PDIA3 could induce apoptosis of CRC cells. The results indicated that siRNA-mediated knockdown of PDIA3 was positively correlated with apoptosis of SW480 cells.
Limitations
There were still some limitations in our study. We examined PDIA3 expression in CRC tissue of patients with TNM-III CRC. It is still unknown whether PDIA3 expression regulates tumor node metastasis. In our study, we detected apoptosis of CRCs after PDIA3 knockdown. However, the specific mechanisms still require clarification.
Conclusion
PDIA3 expression inhibition with siRNA results in subcellular morphological change, inhibition of cellular proliferation and apoptosis. Our study suggests that PDIA3 inhibition could prohibit CRC, likely through inducing apoptosis. PDIA3 would be a potential treatment target for CRC.
Acknowledgments
This work was supported by the Science and Technology Development Project of Jilin Province (20140311092YY), National Natural Science Foundation of China (81572082) and Jilin Provincial Science and Technology Agency Project (2011713, 20150414015GH and 20150101153JC). In addition, the authors would like to thank the Tissue Specimen Bank of China-Japan Union Hospital of Jilin University for supporting this study.
Disclosure
The authors report no conflicts of interest in this work.
References
Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59(4):225–249. | ||
Zheng R, Zeng H, Zhang S, Chen W. Estimates of cancer incidence and mortality in China, 2013. Chin J Cancer. 2017;36(1):66. | ||
Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. | ||
Zhang Z, He Y, Tu X, et al. Mapping of DNA hypermethylation and hypomethylation induced by Folate deficiency in sporadic colorectal cancer and clinical implication analysis of hypermethylation pattern in CBS promoter. Clin Lab. 2017;63(4):733–748. | ||
Loree JM, Kopetz S. Recent developments in the treatment of metastatic colorectal cancer. Ther Adv Med Oncol. 2017;9(8):551–564. | ||
Zhang Y, Baig E, Williams DB. Functions of ERp57 in the folding and assembly of major histocompatibility complex class I molecules. J Biol Chem. 2006;281(21):14622–14631. | ||
Garbi N, Hammerling G, Tanaka S. Interaction of ERp57 and tapasin in the generation of MHC class I-peptide complexes. Curr Opin Immunol. 2007;19(1):99–105. | ||
Garbi N, Tanaka S, Momburg F, Hammerling GJ. Impaired assembly of the major histocompatibility complex class I peptide-loading complex in mice deficient in the oxidoreductase ERp57. Nature Immunol. 2006;7(1):93–102. | ||
Holbrook LM, Sasikumar P, Stanley RG, Simmonds AD, Bicknell AB, Gibbins JM. The platelet-surface thiol isomerase enzyme ERp57 modulates platelet function. J Thromb Haemost. 2012;10(2):278–288. | ||
Grillo C, D’Ambrosio C, Scaloni A, et al. Cooperative activity of Ref-1/APE and ERp57 in reductive activation of transcription factors. Free Radic Biol Med. 2006;41(7):1113–1123. | ||
Zhang J, Wu J, Huo R, et al. ERp57 is a potential biomarker for human fertilization capability. Mol Human Reprod. 2007;13(9):633–639. | ||
Nemere I. The 1,25D3-MARRS protein: contribution to steroid stimulated calcium uptake in chicks and rats. Steroids. 2005;70(5–7):455–457. | ||
Cicchillitti L, Di Michele M, Urbani A, et al. Comparative proteomic analysis of paclitaxel sensitive A2780 epithelial ovarian cancer cell line and its resistant counterpart A2780TC1 by 2D-DIGE: the role of ERp57. J Proteome Res. 2009;8(4):1902–1912. | ||
Coe H, Michalak M. ERp57, a multifunctional endoplasmic reticulum resident oxidoreductase. Int J Biochem Cell Biol. 2010;42(6):796–799. | ||
Chung H, Cho H, Perry C, et al. Downregulation of ERp57 expression is associated with poor prognosis in early-stage cervical cancer. Biomarkers. 2013;18(7):573–579. | ||
Ramos FS, Serino LT, Carvalho CM, et al. PDIA3 and PDIA6 gene expression as an aggressiveness marker in primary ductal breast cancer. Genet Mol Res. 2015;14(2):6960–6967. | ||
Eufemi M, Coppari S, Altieri F, Grillo C, Ferraro A, Turano C. ERp57 is present in STAT3-DNA complexes. Biochem Biophys Res Commun. 2004;323(4):1306–1312. | ||
Liao CJ, Wu TI, Huang YH, et al. Glucose-regulated protein 58 modulates beta-catenin protein stability in a cervical adenocarcinoma cell line. BMC Cancer. 2014;14:555. | ||
Sansone P, Bromberg J. Targeting the interleukin-6/Jak/stat pathway in human malignancies. J Clin Oncol. 2012;30(9):1005–1014. | ||
Anastas JN, Moon RT. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13(1):11–26. | ||
Celli CM, Jaiswal AK. Role of GRP58 in mitomycin C-induced DNA cross-linking. Cancer Res. 2003;63(18):6016–6025. | ||
Chay D, Cho H, Lim BJ, et al. ER-60 (PDIA3) is highly expressed in a newly established serous ovarian cancer cell line, YDOV-139. Int J Oncol. 2010;37(2):399–412. | ||
Herceg Z, Hulla W, Gell D, et al. Disruption of Trrap causes early embryonic lethality and defects in cell cycle progression. Nat Genet. 2001;29(2):206–211. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.