Back to Journals » OncoTargets and Therapy » Volume 11
Expression and functional characterization of FOXM1 in non-small cell lung cancer
Authors Zhang Y, Qiao W, Shan L
Received 14 January 2018
Accepted for publication 7 April 2018
Published 11 June 2018 Volume 2018:11 Pages 3385—3393
DOI https://doi.org/10.2147/OTT.S162523
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Yao Dai
Yan Zhang, Wen-bin Qiao, Li Shan
Department of Thoracic Oncology, Tumor Hospital Affiliated to Xinjiang Medical University, Xinjiang, People’s Republic of China
Objectives: FOXM1 is a key member of the FOX transcription factor family, which plays a vital role in a series of physiological processes. In the present study, non-small cell lung cancer (NSCLC) patients and cell lines were studied to explore the correlation between FOXM1 expression and this malignancy.
Materials and methods: The expression status of FOXM1 was detected in 128 cases of NSCLC tissues and NSCLC cell lines. The relationship of FOXM1 expression and clinicopathological features of NSCLC patients was evaluated by us. In addition, we also explored the biological functions of FOXM1 in NSCLC cell lines.
Results: The FOXM1 is highly expressed in NSCLC tissues and cell lines. FOXM1 expression was closely correlated with lymph node status and TNM stage. Cox regression analysis were performed to demonstrate the prognosis role of FOXM1.
Conclusion: FOXM1 conferred a proliferation and invasion advantage to NSCLC cell. The FOXM1 can be regarded as an important molecular marker in NSCLC prognosis.
Keywords: non-small cell lung cancer, FOXM1, proliferation, prognosis
Introduction
Non-small cell lung cancer (NSCLC) is associated with the highest mortality of any malignancy worldwide. Moreover, morbidity related to NSCLC, for which surgery is the most effective treatment, has been increasing in recent years.1 Early-stage NSCLC is difficult to detect, and the 5-year survival rate among patients with advanced NSCLC is <15%.2 Therefore, it is of vital importance to study the pathogenesis of this disease and identify potential therapeutic targets.
Because cancer progression is a complex process, its pathogenic mechanism remains somewhat elusive.3 Forkhead proteins constitute a class of transcription factors that contain the winged-helix DNA-binding domain.3 Members of the forkhead family play crucial roles in mammalian organ development and cell differentiation and proliferation.4,5 In the present study, NSCLC patients and cell lines were studied to explore the correlation between FOXM1 expression and clinicopathological features of this malignancy, so as to establish the role of FOXM1 in NSCLC development.
Materials and methods
Patients
All specimens derived from 128 NSCLC patients having undergone surgical resection at the Tumor Hospital Affiliated to Xinjiang Medical University between 2011 and 2014. All pathological specimens were fixed in 10% formalin and embedded in paraffin. Histological classification and grading were conducted in accordance with the 2004 World Health Organization (WHO) Histological Classification of Pulmonary Malignant Epithelial Neoplasms, and tumor-node-metastasis (TNM) staging was performed according to the International Association for the Study of Lung Cancer Classification (version 7, 2009).6 The patients had not received pre- or postoperative adjuvant chemotherapy or radiotherapy, and had complete clinicopathological data. Of the 128 patients, 85 were men and 43 were women, and their median age was 61 years (range: 33–73 years). Specimen collection was approved by the Ethics Committee of the Tumor Hospital Affiliated to Xinjiang Medical University and written informed consent was obtained from all patients.
Immunohistochemical staining and evaluation
Immunohistochemistry was conducted using the streptavidin-peroxidase method. The paraffin-embedded specimens were sliced into 4-μm sections before being baked, dewaxed, hydrated, and subjected to antigen retrieval following standard methods. Hydrogen peroxide (3%) was then used to block endogenous peroxidase activity. All procedures were conducted in strict accordance with the instructions provided with the kit used. The sections were then washed thoroughly with phosphate buffered saline (PBS) and incubated with a rabbit anti-human FOXM1 polyclonal antibody (1:100, Proteintech Group, Inc., Rosemont, IL, USA). They were subsequently exposed to a secondary antibody (Zhongshan Golden Bridge Biotechnology, Beijing, People’s Republic of China) for 10 min at 37°C and washed three times with PBS. The sections were then incubated with horseradish peroxidase-conjugated streptavidin for 10 min at 37°C before being washed a further three times with PBS. Later, 3,3′-diaminobenzidine was applied for color development, the degree of which was controlled under a microscope. Finally, distilled water was used to terminate staining.
Five high-power fields were selected for each specimen and comprehensively evaluated, being scored for cytoplasm staining intensity (no staining: 0 points; faint yellow: 1 point; yellow: 2 points; and brown: 3 points) and the percentage of positively stained cells (positive cells <1%: 0 points; 1%–10%: 1 point; 11%–50%: 2 points; and >50%: 3 points). A specimen was deemed positive for FOXM1 expression if the product of its staining intensity and stained cell percentage scores was ≥4 points, and negative if the product of these scores was 0–3 points.
Cell culture
Human lung cancer cells of the lines H1299 and H596, and normal human bronchial epithelial (HBE) cells were purchased from the American Type Culture Collection (Manassas, VA, USA). The RPMI-1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) used in this experiment were supplemented with 10% inactivated fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin.
Western blotting of FOXM1 protein
RIPA lysis solution and phenylmethylsulfonyl fluoride solution were added to cells, which were then centrifuged at 12,000 rpm for 15 min at 4°C. The extracted total protein was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto a polyvinylidene difluoride membrane, which was subsequently immersed in 5% skim-milk blocking solution for 2 h. The membrane was then incubated with a rabbit anti-human FOXM1 polyclonal antibody and mouse anti-human GAPDH monoclonal antibody (1:1,000; Santa Cruz Biotechnologies, Dallas, TX, USA) separately at 4°C overnight, before being exposed to a secondary antibody at 25°C for 1 h. Finally, chemiluminescence reagent was added. Relative FOXM1 expression was calculated as the ratio of FOXM1 to GAPDH gray levels, which were analyzed using Quantity One software (Bio-Rad Laboratories, Hercules, CA, USA).
Enzyme-linked immunosorbent assay (ELISA)
Supernatants from cultures of H1299, H596, and HBE cells were collected and tested using ELISA. FOXM1 in cell lysates was also quantitatively measured with a sandwich ELISA kit for human FOXM1 (JK-Elisa-13146, Jing Kang Bio, Shanghai, People’s Republic of China) according to the manufacturer’s instructions.
Overexpression and RNA interference construct transfection
Cells at the logarithmic phase were plated on a 6-well plate (Corning Incorporated, Corning, NY, USA) at a density of 1×106 cells/well. Cells were transfected upon reaching 50% confluency, washed three times with PBS, and suspended in 200 μL serum-free and double-antibody-free medium. Liposome-mediated transfection was conducted using Lipofectamine 2000 (Thermo Fisher Scientific) in strict accordance with the manufacturer’s instructions. The small interfering RNA (siRNA) sequences were as follows: siFOXM1 forward, 5′-GCCGGAACAUGACCAUCAATT-3′ and reverse, 5′-UUGAUGGUCAUGUUCCGGCGG-3′; scrambled control siRNA forward, 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse, 5′-ACGUGACACGUUCGGAGAATT-3′. Full-length FOXM1 complementary DNA was cloned into the pcDNA3.0 vector (Genechem Co., Ltd., Shanghai, People’s Republic of China) to construct the overexpression plasmid. The empty pcDNA3.0 vector served as the control treatment, and untreated cells were tested in addition to the blank control group. Transfection efficiency was verified by Western blotting and ELISA after 48 h of transfection. Successfully transfected monoclonal cells were selected and amplified for subsequent experiments. Thus, the cells in this study comprised five groups: blank control, siFOXM1, scrambled siRNA, pcDNA-FOXM1, and empty pcDNA3.0.
Cell viability
Cell proliferation was determined using the MTT method. Each group of cells was plated on a 96-well plate and cultured for 48 h, after which, 20 μL MTT solution (5 mg/mL) was added to each well. The plate was then transferred to a 37°C incubator, where it was left for 4 h. The supernatant in each well was discarded upon completion of the reaction. Dimethyl sulfoxide (150 μL) was added to each well and incubated with cells for 10 min at 25°C. The plate was then oscillated for 20 min to ensure thorough mixing and complete dissolution of crystals. The solution in each well was subsequently transferred to a fresh 96-well plate and labeled. The OD of these solutions at a wavelength of 490 nm was measured using a universal microplate spectrophotometer. The OD values of five wells were measured for each group to calculate an average. Finally, cell growth curves were generated.
Cell invasion
Cell invasiveness was tested using a Transwell assay. Cells transfected for 48 h were suspended in FBS-free medium and their density was adjusted to 2×105 cells/mL. This suspension (100–150 μL) was placed in each upper Transwell chamber, and RPMI-1640 medium containing 10% FBS was added to the lower chambers. After 24 h, cells having migrated onto the lower surface of the membrane were collected, fixed with methyl alcohol, and stained with crystal violet. Subsequently, five fields were selected randomly under a microscope and images were taken. Cells were counted and the average cell count was calculated. Triplicate wells were used for each group of cells, and this experiment was repeated three times.
Statistical analysis
Relationships between FOXM1 expression and clinicopathological features were analyzed using the chi-square test. Overall survival (OS) curves were generated using the Kaplan–Meier method, and differences between them were examined using log-rank tests. Cox proportional hazards regression analysis was applied in order to estimate univariate and multivariate hazard ratios for OS. All P-values were two-sided, and those <0.05 were considered statistically significant. All statistical analyses were carried out with SPSS 20.0 software (IBM Corp., Armonk, NY, USA).
Results
Relationship between FOXM1 expression and clinicopathological features of NSCLC patients
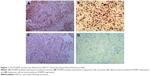
Immunohistochemistry indicated that the FOXM1 protein was mainly expressed in the cytoplasm of NSCLC cells (Figure 1). Immunohistochemical analysis was also used to divide the 128 NSCLC patients into FOXM1-positive (n=90) and FOXM1-negative expression groups (n=38) to establish the relationship between their clinicopathological characteristics and expression of this protein. We found that positive FOXM1 expression was closely correlated with lymph node status and TNM stage (P<0.05), but not with other factors, such as sex, age, smoking history, tumor size, pathological type, and differentiation degree (P>0.05). These data are summarized in Table 1.
  | Table 1 The correlations between FOXM1 expression and clinicopathological features in NSCLC patients |
Relationship between FOXM1 expression and NSCLC patient prognosis
To explore the association between FOXM1 expression and NSCLC prognosis, long-term follow-up was carried out for all of the patients in this study. Follow-up periods ranged from 2 to 85 months, with a median of 42 months. Data concerning the survival of the NSCLC patients are summarized in Table 2. Upon completion of follow-up, survival curves were generated for the 128 patients. Kaplan–Meier analysis indicated that the survival period of NSCLC patients in the FOXM1-positive group was remarkably shorter than that of those in the FOXM1-negative group (Figure 2). In addition, poor differentiation, lymph node metastasis, and advanced TNM stage were associated with worse prognosis. All variables affecting the OS of NSCLC patients were analyzed using the Cox proportional hazards model. Univariate analysis suggested that NSCLC patient prognosis was closely correlated with FOXM1 expression (Table 3). Moreover, multivariate analysis indicated that FOXM1 was an independent predictive factor for patient prognosis. Thus, degree of tumor differentiation, lymph node metastasis, TNM stage, and FOXM1 expression were remarkably correlated with the OS of NSCLC patients.
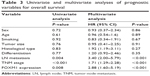  | Table 3 Univariate and multivariate analyses of prognostic variables for overall survival |
FOXM1 expression in NSCLC cell lines and its effect on cell proliferation and invasion
Western blotting indicated that FOXM1 protein is expressed in H1299 and H596 NSCLC cells, and at levels notably higher than those in HBE cells (Figure 3A, left). An ELISA returned similar results (Figure 3A, right). Consequently, we overexpressed and knocked down FOXM1 in H1299 and H596 cells, and confirmed overexpression and siRNA transfection efficiency by Western blotting (Figure 3B) and ELISA (Figure 3C). An MTT assay revealed that growth of FOXM1 siRNA-transfected H1299 and H596 cells was reduced compared to scrambled control siRNA-transfected cells, and FOXM1 overexpression enhanced proliferation (Figure 3D). Furthermore, FOXM1 siRNA-transfected cells exhibited diminished invasiveness compared to control cells, whereas FOXM1 overexpression promoted invasion, as demonstrated by the Transwell assay (Figure 3E).
Discussion
Lung cancer is a leading cause of cancer-related death. According to the WHO, this disease has become the most common malignancy worldwide, being responsible for ~1.6 million deaths annually.7 Lung cancers, 85% of which are NSCLC, mainly originate in the bronchial epithelium. Despite the availability of increasingly advanced medical technologies in recent years, the prognosis of lung cancer has not markedly improved, and the 5-year survival rate remains below 15%.8 Even among patients having received radical surgery at an early stage, the recurrence rate is extremely high. Therefore, there is an urgent need to explore more efficient targeted therapeutics to further improve lung cancer prognosis. Here, we aimed to examine the importance of abnormal FOXM1 expression in the proliferation, invasion, and migration of NSCLC cells.
FOXM1 is a key member of the FOX transcription factor family, which plays a vital role in a series of physiological processes, including regulation of cell cycle-specific genes, cell proliferation and differentiation, and organ development.9–11 Recent research has indicated that FOXM1 can stimulate cell proliferation, which is of great significance to the genesis and development of malignant tumors.12–16 Moreover, many investigations have demonstrated that FOXM1 expression is heightened in multiple solid malignant tumor tissues, including cancers of the prostate, cervix, stomach, and pancreas and NSCLC.17–22 FOXM1 is a pivotal regulator of mitosis. Excessive upregulation of FOXM1 can stimulate gene transcription, ultimately resulting in oncogenesis and malignant tumor progression. In addition, upregulated FOXM1 expression can inhibit apoptosis of lung cancer cells and promote their proliferation. It has been demonstrated in in vitro experiments that suppression of FOXM1 expression by RNA interference or drugs induces cell cycle arrest, restricting proliferation, and can also remarkably increase the rate of apoptosis.23 Upregulated FOXM1 expression and enhanced FOXM1 activity facilitate penetration of the basement membrane by malignant tumor cells, which can then spread to other tissues.24 The above studies have indicated the important role of FOXM1 in the proliferation, invasion, and metastasis of malignant cells, factors that affect cancer patient prognosis.
Elevated FOXM1 expression can be found in both adenocarcinoma and squamous cell carcinoma lung tumor tissues. Furthermore, FOXM1 expression is closely correlated with clinicopathological features such as lymph node metastasis and TNM stage, suggesting that this protein may promote the growth, proliferation, and invasion of NSCLC cells. This view is supported by our present in vitro experiments. Long-term follow-up of the 128 NSCLC patients in this investigation revealed that those expressing FOXM1 at a higher level survived for a shorter time than those exhibiting lower FOXM1 expression. The results of our multivariate regression analysis suggested that high FOXM1 expression is an important independent predictor of poorer prognosis for NSCLC patients. Based on the current study, we can speculate that FOXM1 expression is closely related to the genesis and development of NSCLC, and that the malignant grade of such cancers may positively correlate with the level of this protein. FOXM1 over expression not only promotes tumor formation, but also increases the clinical staging of malignant tumors, the scope of invasion increased lymphatic metastasis will directly affect the prognosis of patients. FOXM1 overexpression inhibited cell apoptosis caused by miR-29.25 Therefore, the examination of FOXM1 expression in malignant tumors can provide important evidences for the clinical diagnosis, treatment and prognosis of malignant tumors. Many findings suggest a role of FOXM1 as an oncogene in the development of NSCLC and in the progression of malignancy. FOXM1 is a regulator of mitosis that can stimulate gene transcription when abnormally up-regulated in its expression level, eventually leading to malignant tumors. FOXM1 is also a proliferation-specific factor whose transcriptional activity is controlled synergistically by multiple signaling pathways and multiple protein factors in biological organisms. High FOXM1 expression in malignant tumor cell lines, including NSCLC, has been reported. Therefore, FOXM1 and NSCLC are closely related to the process of malignant transformation. In addition, overexpression of FOXM1 could increase the migratory and invasive abilities of NSCLC cells. The FOXM1 expression is critical for the invasiveness of malignant NSCLC cells, and this effect was at least in part through epithelial–mechenmychal transition.26 FOXM1 plays an important role in the production of pulmonary blood vessels and the proliferation, invasion, and metastasis of tumor cells, and may also affect the prognosis of NSCLC patients.
Here, we confirmed that FOXM1 is highly expressed in NSCLC tissue. Importantly, we also found that it may constitute a promising biomarker of high risk of invasion and metastasis and poor prognosis in this disease. Regulation of the variety of genes related to and including FOXM1 may be complicated, with relationships between these genes being not simply direct one-to-one interactions, but forming a more complex regulatory network.
Conclusion
The FOXM1 is highly expressed in NSCLC tissues and cell lines. FOXM1 expression was closely correlated with lymph node status and TNM stage. Cox regression analysis was performed to demonstrate the prognosis role of FOXM1. FOXM1 conferred a proliferation and invasion advantage to NSCLC cells. The FOXM1 can be regarded as an important molecular marker in NSCLC prognosis.
Acknowledgment
This project was supported by grants from the Natural Science Foundation of Xinjiang (No 2016D01C378).
Disclosure
The authors report no conflicts of interest in this work.
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. | ||
Akamatsu H, Mori K, Naito T, et al. Progression-free survival at 2 years is a reliable surrogate marker for the 5-year survival rate in patients with locally advanced non-small cell lung cancer treated with chemoradiotherapy. BMC Cancer. 2014;14:18. | ||
Xiaokaiti Y, Wu H, Chen Y, et al. EGCG reverses human neutrophil elastase-induced migration in A549 cells by directly binding to HNE and by regulating α1-AT. Sci Rep. 2015;5:11494. | ||
Golson ML, Kaestner KH. Fox transcription factors: from development to disease. Development. 2016;143(24):4558–4570. | ||
Feuerborn A, Srivastava PK, Küffer S, et al. The forkhead factor FoxQ1 influences epithelial differentiation. J Cell Physiol. 2011;226(3):710–719. | ||
Rami-Porta R, Bolejack V, Giroux DJ, et al. The IASLC lung cancer staging project: the new database to inform the eighth edition of the TNM classification of lung cancer. J Thorac Oncol. 2014;9(11):1618–1624. | ||
Zheng R, Zeng H, Zuo T, et al. Lung cancer incidence and mortality in China, 2011. Thorac Cancer. 2016;7(1):94–99. | ||
Rossi G, Graziano P, Leone A, Migaldi M, Califano R. The role of molecular analyses in the diagnosis and treatment of non-small-cell lung carcinomas. Semin Diagn Pathol. 2013;30(4):298–312. | ||
Leung TW, Lin SS, Tsang AC, et al. Over-expression of FoxM1 stimulates cyclin B1 expression. FEBS Lett. 2001;507(1):59–66. | ||
Katoh M, Katoh M. Human FOX gene family (Review). Int J Oncol. 2004;25(5):1495–1500. | ||
Zhu H. Forkhead box transcription factors in embryonic heart development and congenital heart disease. Life Sci. 2016;144:194–201. | ||
Halasi M, Gartel AL. FOX(M1) news – it is cancer. Mol Cancer Ther. 2013;12(3):245–254. | ||
Gartel AL. FOXM1 in cancer: interactions and vulnerabilities. Cancer Res. 2017;77(12):3135–3139. | ||
Wonsey DR, Follettie MT. Loss of the forkhead transcription factor FoxM1 causes centrosome amplification and mitotic catastrophe. Cancer Res. 2005;65(12):5181–5189. | ||
Laoukili J, Stahl M, Medema RH. FoxM1: at the crossroads of ageing and cancer. Biochim Biophys Acta. 2007;1775(1):92–102. | ||
Wierstra I, Alves J. FOXM1, a typical proliferation-associated transcription factor. Biol Chem. 2007;388(12):1257–1274. | ||
Lokody I. FOXM1 and CENPF: co-pilots driving prostate cancer. Nat Rev Cancer. 2014;14(7):450–451. | ||
He SY, Shen HW, Xu L, et al. FOXM1 promotes tumor cell invasion and correlates with poor prognosis in early-stage cervical cancer. Gynecol Oncol. 2012;127(3):601–610. | ||
Jiang D, Jiang L, Liu B, et al. Clinicopathological and prognostic significance of FoxM1 in gastric cancer: a meta-analysis. Int J Surg. 2017;48:38–44. | ||
Kong FF, Qu ZQ, Yuan HH, et al. Overexpression of FOXM1 is associated with EMT and is a predictor of poor prognosis in non-small cell lung cancer. Oncol Rep. 2014;31(6):2660–2668. | ||
Sun Q, Dong M, Chen Y, Zhang J, Qiao J, Guo X. Prognostic significance of FoxM1 expression in non-small cell lung cancer. J Thorac Dis. 2016;8(6):1269–1273. | ||
Dai J, Yang L, Wang J, Xiao Y, Ruan Q. Prognostic value of FOXM1 in patients with malignant solid tumor: a meta-analysis and system review. Dis Markers. 2015;2015:352478. | ||
Ning Z, Wang A, Liang J, et al. USP22 promotes the G1/S phase transition by upregulating FoxM1 expression via β-catenin nuclear localization and is associated with poor prognosis in stage II pancreatic ductal adenocarcinoma. Int J Oncol. 2014;45(4):1594–1608. | ||
Jin H, Li XJ, Park MH, Kim SM. FOXM1-mediated downregulation of uPA and MMP9 by 3,3′-diindolylmethane inhibits migration and invasion of human colorectal cancer cells. Oncol Rep. 2015;33(6):3171–3177. | ||
Wang X, Zhong H, Wang L, et al. MiR-29 induces K562 cell apoptosis by down-regulating FOXM1. Med Sci Monit. 2015;21:3115–3120. | ||
Xu N, Jia D, Chen W, et al. FoxM1 is associated with poor prognosis of non-small cell lung cancer patients through promoting tumor metastasis. PLoS One. 2013;8(3):e59412. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.