Back to Journals » OncoTargets and Therapy » Volume 12
Expression and Effects of Long Non-Coding RNA, LINC01124, in Non-Small Cell Lung Cancer
Authors Wang ZB, Zhang HY, Lu JB
Received 30 April 2019
Accepted for publication 23 November 2019
Published 3 January 2020 Volume 2019:12 Pages 11729—11736
DOI https://doi.org/10.2147/OTT.S214049
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Arseniy Yuzhalin
Zi-Bo Wang, Hong-Yan Zhang, Ji-Bin Lu
Department of Thoracic Surgery, Shengjing Hospital of China Medical University, Shenyang 110000, People’s Republic of China
Correspondence: Ji-Bin Lu
Department of Thoracic Surgery, Shengjing Hospital of China Medical University, Shenyang 110000, People’s Republic of China
Tel +86 18940251178
Email [email protected]
Objective: To investigate the expression and evaluate the clinical significance of long non-coding RNA, LINC01124, in non-small cell lung cancer (NSCLC) and to study its influence in this tumor.
Methods: Hundred specimens of NSCLC tissues and normal lung tissues after surgery were collected. The qRT-PCR for LINC01124 expression was performed on cancerous and normal lung tissues. The correlations between the expression of LINC01124 and pathological characteristics were analyzed. PcDNA-LINC01124 was transfected to upregulate LINC01124 expression in NSCLC cells, and the transfection efficiency was evaluated by the qRT-PCR. CCK8 assay, wound-healing assay, and the Transwell assay were performed to evaluate the effect of ectopic LINC01124 expression on proliferation, migration, and invasive of NSCLC cells.
Results: The expression level of LINC01124 was downregulated in tumor tissues when compared with the paired normal lung tissues (P<0.05). The expression of LINC01124 was associated with patients’ age and distant metastasis (P<0.05). Enforced expression of LINC01124 significantly inhibited the proliferation, migration, and invasive ability of NSCLC cells.
Conclusion: The expression of LINC01124 was decreased in patients with NSCLC of older age and with those having distant metastasis. LINC01124 may inhibit cell proliferation, migration, and invasive ability.
Keywords: non-small cell lung cancer, NSCLC, long non-coding RNA, lncRNA, LINC01124
Introduction
Lung cancer is one of the most fatal malignant tumors, with the number of new cases worldwide exceeding 1.8 million per year.1 In 2015, there were approximately 4,292,000 new cases of cancer in China and 2.8 million associated deaths, among which lung cancer was one of the leading causes.2 Many pathological types of lung cancer exist, but approximately 85% of lung cancer patients are diagnosed with non-small cell lung cancer (NSCLC), which has a 5-year survival rate of only 10%.3 Although there has been significant progress made in recent years in improving diagnoses and lung cancer treatments, the prognoses remain unoptimistic. Therefore, further research into the molecular mechanisms behind the development and progression of NSCLC is imperative.4,5
The human genome contains approximately 20,000 protein-coding genes, which account for only 2% of the total genes. More than 90% of transcripts are non-coding RNAs. Long non-coding RNAs (lncRNAs) are a type of non-coding RNA, greater than 200 nucleotides in length.6,7 They have no open reading frame, and cannot be translated into protein. Increasing evidence suggests that lncRNAs play indispensable roles in the proliferation, growth, and apoptosis of tumor cells.8 The lncRNAs participate in the development and progression of tumors by interfering with processes related to tumorigenesis.9 Studies have confirmed that abnormal lncRNA expression is associated with a variety of cancers, including lung cancer, although the specific mechanisms by which lncRNAs influence tumorigenesis remain unclear. The lncRNAs play a dual role in lung cancer, acting as both oncogene and tumor suppressor gene.10,11 Therefore, in the vast network of cancer-related processes, it is particularly important to explore the role of lncRNAs in tumor formation and metastasis.
Long intergenic non-protein coding RNA 1124 (LINC01124) is located on chromosome 2q31.1, and contains one exon. In this study, we investigated the expression of LINC01124 in cancer tissue and adjacent normal tissue samples from 100 patients with NSCLC, and analyzed correlations between expression and clinical features of NSCLC. By adopting this approach, we explored whether LINC01124 had the potential to be a new biomarker for the prediction and diagnosis of metastatic NSCLC. Finally, we investigated the effects of LINC01124 on the proliferation, migration, and invasion of lung cancer cells using several in vitro assays.
Methods
Patients and Tissue Samples
Hundred NSCLC tissues and matched adjacent non-tumor tissues were collected from patients in Shengjing Hospital between 2016 and 2017. All patients recruited in this study were not subjected to preoperative radiotherapy or chemotherapy. This research was approved by the Ethics Committee of Shengjing Hospital, and human samples were obtained with written informed consent from all patients. All tissue samples were excised and stored in liquid nitrogen immediately until total RNA extraction.
Total RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Analysis
Total RNA was extracted from frozen tissue samples or cells using TRIzol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocols. For reverse transcription, 500 ng total RNA was reverse transcribed in a final volume of 20 μL using the PrimerScript RT reagent Kit (TaKaRa, Beijing, China) according to the manufacturer protocol. The qRT-PCR was performed using the SYBR Premix Ex TaqⅡ(TaKaRa, Beijing, China) on 7900HT Fast Real-Time PCR system according to the manufacturer’s instructions. The primers are shown in Table 1. Ct-values were calculated using SDS 2.4 software. GAPDH was used as an internal control. The Ct-value for each sample was calculated with the ΔΔCt-method, and the results were expressed as 2-ΔΔCT to analyze the fold change (tumor vs normal): ΔΔCT=(CTtarget gene–CTactin)normal–(CTtarget gene–tCTactin)tumor.
 |
Table 1 Sequences of Primers for qRT-PCR |
Cell Culture
A549 cell line and LTE cell line were purchased from the Institute of Biochemistry and Cell Biology of Chinese Academy of Science (Shanghai, China). Cells were incubated at 37.5°C with 5% CO2 in RPMI 1640 medium supplemented with 10% fetal bovine serum (10% FBS, GIBCO), 100 U/mL penicillin, and 100 mg/mL streptomycin.
Overexpression Vector Transfection
We used a Linc01124-overexpression vector (pcDNA3.1-linc01124) (Nanjing GenScript Biotech Corp, Nanjing, China) to induce overexpression of linc01124. The A549 and LTE cells were transfected by pcDNA3.1-linc01124 for 48 hrs and the overexpression efficiency was detected by qPCR.
Cell Proliferation Assay
Cell proliferation was determined using the cell counting kit-8 (CCK-8) assay (Dojindo, Japan, Tokyo), according to the manufacturer’s instructions. After transfection, A549 cells and LTE cells were seeded into 96-well plates (2×104 cells/well), respectively, and incubated in RPMI 1640 at 37°C and 5% CO2 atmosphere. At 24, 48, and 72 hrs, CCK-8 (10 μL/well) solution was added to measure cell viability. After 1.5 hrs, the absorbance of each well was measured at 450 nm. Each experiment was repeated at least three times independently.
Wound-Healing Assay
Wound-healing assay was performed to detect the migration ability and compare the difference among the two experimental groups (pcDNA3.1-linc01124 and blank pcDNA3.1) of A549 and LTE cells, respectively. 2×105 cells/well were seeded in 6-well plates. When cells were completely adherent, a sterile 20 μL tip was used to scratch a straight line through the cell layer in each well. Cell migration was determined by detecting the average distance the growing cells had migrated into the wound surface under microscopy at the designated time periods. Assays were repeated three times for each clone.
Transwell Assay
The Transwell assay was performed to detect the migration and invasive abilities. A549 and LTE cells were grown in RPMI 1640 medium containing 10% FBS and transfected with pcDNA3.1-linc01124 and the blank pcDNA3.1. After 24 hrs, the cells were harvested by trypsinization and washed once with phosphate-buffered saline.
To measure cell migration, Transwell chambers (8 μm; Corning Incorporated, Corning, NY, USA) were placed into the wells of 24-well culture plates. Matrigel transwell chambers were used to measure cell invasion.
A total of 1×105 cells were cultured in 100 μL of serum-free RPMI 1640 medium added into the upper chamber of the Transwells. In the lower chamber, 600 μL of RPMI 1640 containing 20% FBS was added. After 24 hrs of incubation at 37°C with 5% CO2, the migrated cells were stained with DAPI (Beyotime, China, Shanghai). The number of invaded cells was counted and the average value was determined through three different random fields.
Results
Detection of LINC01124 Expression by qRT-PCR
The qRT-PCR was used to detect the expression levels of LINC01124 in cancer tissue and adjacent normal tissue from NSCLC samples. Results showed that the LINC01124 expression levels in lung cancer tissue were significantly lower than normal tissue (t=−19.969, P<0.01) (Figure 1).
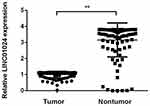 |
Figure 1 The LINC01124 expression levels in lung cancer tissue and normal lung tissues. **P<0.01. |
The recombinant plasmid pcDNA3.1-LINC01124 and blank-plasmid pcDNA3.1 were transfected into both lung cancer A549 and LTE cells, and LINC01124 expression was detected. Each experiment was repeated at least three times. At 48 hrs after transfection, transfection efficiency was determined using qRT-PCR. As depicted in Figure 2 (A and B), the transfected pcDNA3.1-LINC01124 significantly increased LINC01124 expression in both A549 and LTE cells. We used pcDNA3.1-LINC01124 to perform further analysis.
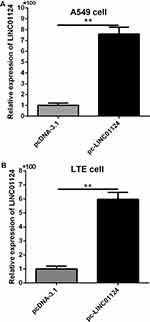 |
Figure 2 Plasmids and pc-linc01124 were, respectively, transfected into A549 (A) and LTE cells (B) to obtain LINC01124 overexpression. **P<0.01. |
Correlation Between LINC01124 Expression and Clinical Features of NSCLC
Using the median value of LINC01124 expression levels in cancer tissue as a cutoff, our 100 patient cohort was divided into a high expression group (n=50) and a low expression group (n=50). Correlations between LINC01124 expression levels and clinical features were analyzed using a chi-squared test. We found that LINC01124 expression levels showed significant correlations with age and distant metastasis (P<0.05); older patient age correlated with lower LINC01124 expression levels, and patients with distant metastasis had further lower LINC01124 expression levels. There were no significant correlations between LINC01124 expression and sex, smoking status, lymph node metastasis, tumor size, or tumor differentiation (P>0.05) (Table 2).
 |
Table 2 Correlation Between Clinicopathological Features and LINC01024 Expression in NSCLC Patients |
The Effects of LINC01124 Expression on Cellular Proliferation
A CCK-8 assay revealed that LINC01124 expression in A549 and LTE cells was increased following transfection of pcDNA3.1-LINC01124. When compared with the control group (blank plasmid), the proliferation of LINC01124-overexpressing A549 (Figure 3A) and LTE (Figure 3B) cells was significantly inhibited.
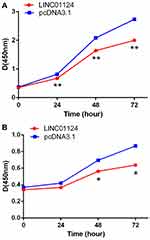 |
Figure 3 The proliferation ability of LINC01124-overexpressing cells and blank cells (A) A549 cells (B) LTE cells. *P<0.05, **P<0.01. |
The Effects of LINC01124 Expression on Cellular Migration
To investigate the influence of LINC01124 on migration in NSCLC cell lines, we performed a wound-healing assay. The number of cells in the scratched area in the pcDNA3.1-LINC01124 group was significantly less compared with the control group (blank plasmid) at the designated time periods (24 hrs, 48 hrs). The assay showed that overexpression of LINC01124 leads to prolonged scratch wound closure in A549 and LTE cells as compared with control cells (Figure 4A, B).
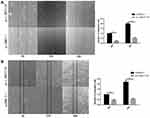 |
Figure 4 The results of wound-healing assay in LINC01124-overexpressing cells and blank cells. (A) A549 cells (B) LTE cells. **P<0.01. |
Transwell cell invasion assay indicated that the ability of LINC01124-overexpressed A549 cells and LTE cells to migrate in the transwell chamber significantly decreased compared with empty vector controls (Figure 5A, B).
The Effects of LINC01124 Expression on Cellular Invasion
We next performed a Transwell assay with Matrigel, which demonstrated that A549 and LTE cell invasion is inhibited after LINC01124 overexpression as compared with empty vector controls (Figure 5A, B).
Discussion
NSCLC is one of the leading causes of cancer-related deaths worldwide.12,13 Due to the limitations in early diagnosis of NSCLC, its 5-year survival rate is still less than 20%.14 The lncRNAs were once considered to be “transcriptional noise”; however, accumulating evidence suggests that lncRNAs have important biological functions.15–17 Studies have shown that lncRNAs play critical roles in both physiological and pathological cellular processes,18,19 are involved in the development of various diseases including tumors,20 and are associated with lymphatic and distant metastasis, impacting on patient prognosis.21
A number of NSCLC-related lncRNAs have been discovered.22–24 For example, in 2013, Liu et al found that the lncRNA HOTAIR showed significantly elevated expression in NSCLC, and its high expression levels were closely related to lymph node metastases and patient prognoses.25 Zhang et al, in their meta-analysis, found that high expression levels of the lncRNA MALAT1 were associated with lung cancer prognosis.26,27 In our study, we found that the lncRNA LINC01124, located on chromosome 2q31.1, may be involved in NSCLC. First, in examining NSCLC tissues, we found that LINC01124 expression levels were significantly lower than those in adjacent normal tissues. Second, by increasing LINC01124 expression levels in NSCLC cells in vitro, we significantly inhibited the proliferation, migration, and invasion of these cells, demonstrating that LINC01124 suppresses key tumorigenic properties of NSCLC cells.
We first detected the expression of LINC01124 in the lung cancer tissue samples and adjacent normal tissue samples from 100 patients with NSCLC. We found that the expression of LINC01124 was significantly lower in NSCLC tissues than in normal adjacent tissues. We then analyzed the clinical features of 100 patients and found a significant correlation between LINC01124 levels and patient age and distant metastasis.
We further studied the effect of LINC01124 on in vitro cultured NSCLC cells. We transfected the recombinant plasmid pcDNA3.1-LINC01124 into A549 and LTE cells. The proliferation, migration, and invasion of LINC01124-overexpressed A549 and LTE cells significantly decreased, thereby suggesting that LINC01124 suppressed the proliferation, migration, and invasive abilities of NSCLC cells.
Relatively few studies on LINC01124 have been published, and its function in NSCLC and other cancers remains unclear. Our results suggested that LINC01124 had inhibitory effects in NSCLC, although we lacked in vivo data to support our observations; therefore, in vivo studies are warranted in the future to delineate the precise role of this molecule in lung cancer etiology.
In conclusion, our study demonstrated the important role of LINC01124 in the development of NSCLC and regulation of biological functions of cancer cells. It is expected to be a new biomarker and molecular target for the diagnosis and treatment of NSCLC.
Funding
Social development plan of Shenyang in 2013 (130245).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262
2. Yang T, Chen BZ, Li DF, et al. Reduced NM23 protein level correlates with worse clinicopathologic features in colorectal cancers: a meta-analysis of pooled data. Medicine (Baltimore). 2016;95(4):e2589. doi:10.1097/MD.0000000000002589
3. Verdecchia A, Francisci S, Brenner H, et al. Recent cancer survival in Europe: a 2000–02 period analysis of EUROCARE-4 data. Lancet Oncol. 2007;8(9):784–796. doi:10.1016/S1470-2045(07)70246-2
4. Thomson CS, Forman D. Cancer survival in England and the influence of early diagnosis: what can we learn from recent EUROCARE results? Br J Cancer. 2009;101(Suppl 2):S102–S109. doi:10.1038/sj.bjc.6605399
5. Wu Y, Liu H, Shi X, Yao Y, Yang W, Song Y. The long non-coding RNA HNF1A-AS1 regulates proliferation and metastasis in lung adenocarcinoma. Oncotarget. 2015;6(11):9160–9172. doi:10.18632/oncotarget.3247
6. Tantai J, Hu D, Yang Y, Geng J. Combined identification of long non-coding RNA XIST and HIF1A-AS1 in serum as an effective screening for non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8(7):7887–7895.
7. Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839(11):1097–1109. doi:10.1016/j.bbagrm.2014.08.012
8. Zhu J, Fu H, Wu Y, Zheng X. Function of lncRNAs and approaches to lncRNA-protein interactions. Sci China Life Sci. 2013;56(10):876–885. doi:10.1007/s11427-013-4553-6
9. Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38–55. doi:10.1186/1476-4598-10-38
10. Yang Q, Xu E, Dai J, et al. A novel long noncoding RNA AK001796 acts as an oncogene and is involved in cell growth inhibition by resveratrol in lung cancer. Toxicol Appl Pharmacol. 2015;285(2):79–88. doi:10.1016/j.taap.2015.04.003
11. Hu T, Lu YR. BCYRN1, A c-MYC-activated long non-coding RNA, regulates cell metastasis of non-small-cell lung cancer. Cancer Cell Int. 2015;15:36. doi:10.1186/s12935-015-0183-3
12. DeSantis CE, Lin CC, Mariotto AB, et al. Cancer treatment and survivorship statistics. CA Cancer J Clin. 2014;64(4):252–271. doi:10.3322/caac.21235
13. Sanoff HK, Sargent DJ, Campbell ME, et al. Five-year data and prognostic factor analysis of oxaliplatin and irinotecan combinations for advanced colorectal cancer: N9741. J Clin Oncol. 2008;26(35):5721–5727. doi:10.1200/JCO.2008.17.7147
14. De Rosa M, Pace U, Rega D, et al. Genetics, diagnosis and management of colorectal cancer. Oncol Rep. 2015;34(3):1087–1096. doi:10.3892/or.2015.4108
15. Loewer S, Cabili MN, Guttman M, et al. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet. 2010;42(12):1113–1117. doi:10.1038/ng.710
16. Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142(3):409–419. doi:10.1016/j.cell.2010.06.040
17. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi:10.1038/nrg2521
18. Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: new links in cancer progression. Cancer Res. 2011;71(1):3–7. doi:10.1158/0008-5472.CAN-10-2483
19. Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31(43):4577–4587. doi:10.1038/onc.2011.621
20. Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Mod Pathol. 2013;26(2):155–165. doi:10.1038/modpathol.2012.160
21. Li CH, Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. Int J Biochem Cell Biol. 2013;45(8):1895–1910. doi:10.1016/j.biocel.2013.05.030
22. Sung JJ, Lau JY, Young GP, et al. Asia Pacific consensus recommendations for colorectal cancer screening. Gut. 2008;57(8):1166–1176. doi:10.1136/gut.2007.146316
23. Rerknimitr R, Angsuwatcharakon P, Ratanachu-ek T, et al. Asia-Pacific consensus recommendations for endoscopic and interventional management of hilar cholangiocarcinoma. J Gastroenterol Hepatol. 2013;28(4):593–607. doi:10.1111/jgh.2013.28.issue-4
24. Arab K, Park YJ, Lindroth AM, et al. Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol Cell. 2014;55(4):604–614. doi:10.1016/j.molcel.2014.06.031
25. Liu XH, Liu ZL, Sun M, Liu J, Wang ZX, De W. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer. 2013;13:464. doi:10.1186/1471-2407-13-464
26. Zhang J, Zhang B, Wang T, Wang H. LncRNA MALAT1 overexpression is an unfavorable prognostic factor in human cancer: evidence from a meta-analysis. Int J Clin Exp Med. 2015;8(4):5499–5505.
27. Feng J, Sun Y, Zhang EB, Lu XY, Jin SD, Guo RH. A novel long noncoding RNA IRAIN regulates cell proliferation in non small cell lung cancer. Int J Clin Exp Pathol. 2015;8(10):12268–12275.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.