Back to Journals » International Journal of General Medicine » Volume 14
Expression and Clinical Significance of Origin Recognition Complex Subunit 6 in Breast Cancer – A Comprehensive Bioinformatics Analysis
Authors Chen S, Jin Z, Xin L, Lv L, Zhang X, Gong Y, Liu J
Received 15 October 2021
Accepted for publication 18 November 2021
Published 14 December 2021 Volume 2021:14 Pages 9733—9745
DOI https://doi.org/10.2147/IJGM.S342597
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Shaohua Chen,1,2 Ziyao Jin,3 Linfeng Xin,4 Lv Lv,2 Xuemei Zhang,3 Yizhen Gong,5 Jianlun Liu1
1Department of Breast Surgery, Guangxi Medical University Cancer Hospital, Nangning, People’s Republic of China; 2Department of Breast and Thyroid Surgery, Affiliated Hospital of Guilin Medical University, Guilin, People’s Republic of China; 3Department of Pathology, Affiliated Hospital of Guilin Medical University, Guilin, People’s Republic of China; 4Clinical Medicine, Guilin Medical University, Guilin, People’s Republic of China; 5Department of Gastrointestinal Surgery, Guangxi Medical University Cancer Hospital, Nangning, People’s Republic of China
Correspondence: Jianlun Liu Email [email protected]
Objective: We aimed to investigate the expression, diagnostic and prognostic values, and potential molecular mechanisms of the origin recognition complex (ORC) in breast cancer (BC).
Methods: Kaplan–Meier estimation was used to assess the prognostic value of ORC genes, and Oncomine, TCGA, GEO and ULCAN databases were used to analyze their expression in BC. Wilcoxon rank-sum tests were used to evaluate the relationship between ORC gene expression levels and BC clinicopathological features. Receiver operating characteristic (ROC) curves were used to assess the diagnostic value of ORC genes in BC. Survival analysis was performed using Kaplan–Meier estimation and Cox regression. A nomogram was constructed to predict 1-, 3-, and 5-year survival probabilities in BC. Gene set enrichment analysis (GSEA) and immune infiltration were used to investigate potential molecular mechanisms of the ORC.
Results: ORC1L and ORC6L were highly expressed in BC compared with healthy tissue, while ORC5L expression patterns were inconsistent; no significant differences in ORC2L, ORC3L or ORC4L expression were observed between BC and healthy tissues. ORC1L and ORC6L expression levels were significantly correlated with age, tumor (T) stage and molecular subtype; ORC5L expression was significantly correlated with age and number of nearby lymph nodes with cancer (N stage). ORC6L expression had the highest diagnostic value in BC and was an independent prognostic factor for poor overall survival (OS). ORC6L may be involved in cell cycle progression and may regulate cancer signaling pathways, including NF-κB, P53, and WNT, in BC. ORC6L expression was also associated with immune infiltration.
Conclusion: ORC1L and ORC6L are highly expressed in BC; ORC6L has a high diagnostic value and is an independent prognostic factor for poor OS. ORC6L may be involved in the initiation and progression of BC by regulating cell cycle progression, promoting cancer signaling pathway activation, and influencing tumor immune cell infiltration.
Keywords: origin recognition complex, breast cancer, prognostic biomarker, nomogram, gene set enrichment analysis, immune infiltration
Introduction
Breast cancer (BC) is the most common malignancy worldwide. It is the most common cause of cancer-related deaths in women and the fifth leading cause of cancer-related deaths globally.1 Despite rapid developments in diagnostic methods and treatment strategies for this disease in recent decades, the 5-year stage IV BC survival rate is only 20%, and 5–10% of newly diagnosed BC patients have distant metastases.2 Therefore, novel diagnostic and prognostic biomarkers of BC and a better understanding of BC pathogenesis are critical to improving prognosis and survival for people with this cancer type.
Recently, bioinformatics analyses identified two of the six subunits of the origin recognition complex (ORC), ORC1L and ORC6L, are related to the maintenance of the fundamental stem cell properties of triple negative breast cancer (‘stemness’) and cognitive dysfunction in breast cancer survivors, respectively.3,4 The ORC binds to the origin of DNA replication and regulates the initiation of replication.5 In recent decades, substantial evidence has shown that the ORC is involved in other biological processes and conditions, such as heterochromatin formation, cytokinesis, viral replication, DNA damage repair, cognitive impairment, and radiation inflammatory response.6–11 ORC genes are also known to be differentially expressed in a number of tumors, such as gliomas,12 endometrial cancer,13 colon cancer,14–16 gastric cancer,17,18 liver cancer,19 esophageal cancer,20 and lung cancer.21 They are implicated in diverse biological processes, including cell proliferation, migration, invasion, and chemotherapy resistance.13–15,21–23 ORC1L, ORC5L and ORC6L have known prognostic value in gastric cancer, liver cancer, and colorectal cancer;16–19 however, the expression levels and roles of ORC subunits in BC tumorigenesis remain to be elucidated.
In the current study, we aimed to assess the expression levels, clinical values, and potential molecular mechanisms of ORC subunits in BC, providing a new theoretical basis for the diagnosis and treatment of BC.
Materials and Methods
Kaplan–Meier Analysis
We assessed the association between ORC genes and overall survival (OS) in BC patients using the Kaplan–Meier Plotter online tool (http://kmplot.com/analysis/, accessed on: July 16, 2021). This tool is capable of assessing the effects of more than 54,000 genes (mRNA, miRNA, protein) on survival in 21 cancer types including BC (n = 7830). The sources of data in this tool include the GEO, EGA, and TCGA databases.24 P-values of < 0.05 were considered statistically significant in this study.
ORC Gene Expression and Clinical Features of BC
mRNA levels were used as a proxy to assess ORC gene expression in BC and healthy tissues, based on information in the TCGA database (https://portal.gdc.cancer.gov/, accessed on: July 16, 2021), the GEO database (accession numbers GSE54002 and GSE37751; https://www.ncbi.nlm.nih.gov/geo/, accessed on: July 16, 2021), and the Oncomine platform (www.oncomine.org, accessed on: July 16, 2021). We obtained gene expression data and clinical information from 1109 BC and 113 healthy breast tissue samples from the TCGA database. RNAseq data in fragments per kilobase per million mapped reads (FPKM) were converted to transcripts per million mapped reads (TPM) and log2-converted for subsequent analysis. The clinical characteristics of BC are presented in Table 1.
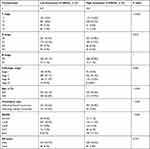 |
Table 1 The Clinical Characteristics of BC Patients (TCGA) |
ORC Protein Expression Analysis
We explored the protein expression of the ORCs in BC in the UALCAN web resource for cancer-omics data analysis (http://ualcan.path.uab.edu/, accessed on: July 16, 2021) using the Clinical Proteomic Tumor Analysis Consortium (CPTAC) confirmatory/discovery dataset.25
Construction and Verification of Nomogram
We used the independent factors found to be associated with BC prognosis in this study to construct a nomogram to predict 1, 3, and 5-year OS. The nomogram was generated in R, using the rms and survival packages.26 Harrell’s concordance index (C-index) was used to quantify the predictive accuracy for each factor, ranging from 0.5 (no predictive power) to 1 (perfect prediction). In addition, calibration plots were generated to examine the performance characteristics of the predictive nomogram.
Gene Set Enrichment Analysis (GSEA)
GSEA (http://www.broadinstitute.org/gsea/index.jsp, accessed on: July 16, 2021) was used to assess the correlation between gene expression signatures based on low versus high ORC6L expression. GSEA is generally used to determine whether a set of genes show statistically significant, concordant differences between two biological states, in this case, BC versus healthy tissue.27 Publicly available microarray expression data for 1109 BC samples were downloaded from TCGA; the significance threshold in our GSEA was set at P < 0.05 and the false discovery rate (FDR) at <0.25.
Assessment of Correlation Between BC Risk and Immune Cell Infiltration
The relative tumor infiltration levels of 24 immune cell types were quantified using ssGSEA of GSVA package.28 The correlation between risk scores and immune infiltration was calculated using Spearman’s rank correlation coefficient analysis. Results with correlation coefficient |r| > 0.3 and P < 0.05 were considered statistically significant.
Statistical Analysis
Statistical analysis and mapping were performed in R (v3.6.3). Wilcoxon rank sum tests were used to compare gene expression between BC and normal breast tissues and to assess the correlation between gene expression and clinicopathological features. Receiver operating characteristic (ROC) curves were plotted using the pROC package. Kaplan–Meier and Cox proportional risk regression models were used to evaluate the prognostic value of ORC genes. P values <0.05 were considered statistically significant.
Results
ORC Gene Expression Levels in BC
We found that the expression of ORC1L and ORC6L were significantly increased and ORC5L significantly decreased in BC compared with healthy tissue (Figure 1A). Furthermore, patients with high levels of ORC5L expression were found to have significantly worse OS (hazard ratio [HR] = 1.22, 95% confidence interval [CI] = 1.01–1.47, Log rank P = 0.036). Similarly, BC patients with high levels of ORC6L expression also had significantly worse OS [HR = 1.5, 95% CI = 1.24–1.81, Log rank P < 0.001], ie, the expression of ORC5L and ORC6L were negatively correlated with OS in people with BC (Figure 1B). In contrast, ORC1L, ORC2L, ORC3L, and ORC4L expression levels were not associated with OS in those with BC. These results suggest that ORC5L and ORC6L may be useful prognostic factors in BC.
To account for tissue-specific expression of ORC genes, we studied their expression in BC a combined multiple database. ORC1L, ORC5L, and ORC6L expression levels were significantly higher in BC than in healthy tissues, while ORC2L, ORC3L, and ORC4L expression levels were not significantly different (Figure 2A). Results of analyses using the GEO datasets GSE54002 and GSE37751 had concordant results (Figure 2B and C). Our comprehensive analysis using the Oncomine, TCGA and GEO databases therefore showed that ORC1L and ORC6L expression was significantly increased in BC compared with healthy tissue, while the results for ORC5L were inconsistent between databases, and there were no significant differences in the expression of ORC2L, ORC3L or ORC4L in BC versus healthy tissue.
UALCAN cancer-omics data were further used to reveal ORC protein levels in 125 BC and 18 normal tissue samples. Consistent with the gene expression results, ORC1L and ORC6L protein levels were significantly increased in BC compared with healthy tissues. However, ORC5L levels were low in BC compared with healthy tissues, which was inconsistent with the gene expression levels (Figure 2D). Interestingly, ORC2L and ORC3L levels were different between BC and healthy tissues despite no significant difference in gene expression in this study, indicating post-transcriptional regulation of these factors in BC. In conclusion, ORC1L, ORC5L and ORC6L were found to be differentially expressed in BC tissues; these proteins may therefore affect incidence and development of BC.
Relationship Between ORC Subunits and Clinicopathological Characteristics in BC
We further explored the relationship between the expression of ORC1L, ORC5L, and ORC6L and the clinicopathological characteristics of BC patients using clinical data from TCGA, including tumor patient age, molecular subtype, tumor volume, lymph node metastasis, distant metastasis, and pathological stage. Our results suggested that high ORC1L and ORC6L expression occurred mainly in the HER2-enriched subtype of BC; their expression was negatively correlated with patient age and positively correlated with tumor size but not with lymph node metastasis, distant metastasis, or tumor stage (Figure 3A and C). ORC5L expression was also negatively correlated with age and positively correlated with lymph node metastasis but not with BC molecular subtype, tumor size, or any other characteristics (Figure 3B).
Diagnostic and Prognostic Values of ORC1L, ORC5L and ORC6L in BC
ROC curves were used to evaluate the diagnostic value of ORC1L, ORC5L, and ORC6L in BC. ORC6L had the highest diagnostic value, with an AUC of 0.907 (95% CI = 0.882–0.932) (Figure 4A). The AUC for ORC1L was 0.885 (95% CI = 0.858–0.913), and the AUC for ORC5L was 0.739 (95% CI = 0.694–0.785).
We used the Kaplan–Meier method to assess the impacts of ORC1L, ORC5L, and ORC6L on patient survival. Interestingly, ORC1L, ORC5L, and ORC6L expression were not significantly correlated with OS in BC patients (Figure 4B). This is inconsistent with previous results that ORC5L and ORC6L have prognostic value in the context of survival based on KM-plot online analysis. Since the expressions of ORC1L, ORC5L, and ORC6L were related to clinicopathological factors, such as age and molecular subtype, univariate and multivariate Cox proportional risk regression models were used to eliminate any confounding effects of these factors. Univariate analysis showed that T stage (size and extent of the tumor), N stage (number of nearby lymph nodes with cancer), M stage (metastatic status), pathological stage, molecular subtype, and age were found to be significantly associated with OS. Further multivariate regression analyses were performed, in which ORC6L level, N stage, M stage, pathological stage, molecular subtype, and age were determined to be independent risk factors in OS (Table 2, Figure 5A). The risk of death in people with high ORC6L expression was found to be 1.538-fold higher than in those with low ORC6L expression.
 |
Table 2 Univariate and Multivariate Cox Regression for OS in BC Patients (TCGA) |
We included all statistically significant factors, including N stage, M stage, pathological stage, molecular subtype, age, and ORC6L expression, in a multivariate Cox proportional risk regression model to construct a nomogram for predicting 1, 3, and 5-year survival in people with BC (Figure 5B). The C-index, calculated by internal re-sampling verification, was 0.716 (95% CI 0.692–0.741), indicating that the predicted results were likely in good agreement with the real results. Our calibration plot also indicated that the nomogram was well calibrated, with mean predicted probabilities close to observed probabilities (Figure 5C). ORC6L therefore has prognostic value in BC.
Enrichment Analysis of ORC6L-Related Signaling Pathways in BC
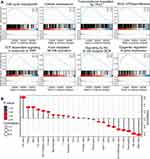
A GSEA was performed to identify the potential molecular mechanisms of ORC6L in BC. In this analysis, patients in the TCGA database were divided into “high” and “low” expression groups based on the median value of ORC6L expression in people with BC in this study. Overall, 14 gene sets were strongly enriched in those with high compared with low ORC6L expression (Figure 6A, Table 3). Some of these gene sets, including those involved in cell cycle checkpoints and cellular senescence, were markedly activated in those with high ORC6L expression, implying a potential cell cycle regulatory mechanism for ORC6L in BC. Pathways that modulate cancer-related functions, including transcriptional regulation by P53, FCER1-mediated nuclear factor (NF-κB) activation, and T-cell factor (TCF)-dependent WNT signaling, were positively correlated with ORC6L expression. In addition, GSEA revealed a relationship between ORC6L expression and activation of Rho GTPase effectors, B cell receptor (BCR) signaling, and epigenetic regulation of gene expression. Together, these results indicate that ORC6L may play an oncogenic role in BC by regulating the cell cycle and activating tumor-related signaling pathways.
 |
Table 3 Signaling Pathways Enriched in the ORC6L High-Expression Group in BC Patients (TCGA) |
Correlation Between ORC6L and Immune Cell Infiltration in BC
Tumor growth and development are determined by both cancer cell-autonomous and microenvironmental mechanisms, including infiltration of the tumor by immune cells. Evidence has shown that tumor-infiltrating lymphocytes play an important role in the development and progression of BC.29,30 Therefore, the ssGSEA of the GSVA package28 was used to determine whether ORC6L modulates the tumor microenvironment. We found that ORC6L was significantly related to the infiltration of various immune cells in BC (Figure 6B). In particular, it was strongly positively correlated with T helper 1 and 2 cell (Th1 and Th2), T regulatory cell (Treg), and dendritic cell (DC) infiltration in BC. ORC6L expression was also closely negatively correlated with mast cell, natural killer (NK) cell, eosinophil, and Th17 cell infiltration in BC. These results suggest that ORC6L may be involved in the occurrence and progression of BC through the regulation of immune cell infiltration.
Discussion
In this study, we demonstrated for the first time that ORC subunits are differentially expressed in people with BC compared with healthy individuals, and that some ORC subunits have diagnostic and prognostic values in this disease. The ORC was previously known to have a role in regulating the initiation of DNA replication, and its role in various tumors has attracted increasing attention in recent years. A previous study showed that ORC1L is upregulated in glioma and promotes the proliferation, migration and invasion of glioma cells by activating the extracellular signal-related kinase (ERK)/c-Jun N-terminal kinase (JNK) signaling pathway.12 In lung cancer and triple-negative BC, ORC1L is considered a pivotal gene in the maintenance of tumor stem cells, and its expression in lung cancer is positively correlated with tumor stage.3,21 ORC5L and ORC6L are known to have prognostic value in hepatocellular carcinoma.19 It was reported that ORC subunits are also associated with chemotherapy resistance in various tumor types: for example, upregulation of ORC2L and ORC5L has been detected in cisplatin-resistant bladder cancer cell lines;23 in pancreatic cancer, ORC4L is involved in phosphoinositide 4-kinase (PIK1)-mediated resistance to gemcitabine;22 and in colorectal cancer cells, high expression of ORC6L has been shown to promote cell proliferation and induce resistance to the chemotherapeutic agents fluorouracil and cisplatin.15 Therefore, our study adds to the evidence that ORCs have potential clinical applications in a variety of tumors, including BC.
We demonstrated that ORC1L and ORC6L mRNA and protein levels were significantly increased in BC compared with healthy tissues in an analysis of data from the Oncomine, TCGA, GEO and ULCAN databases. Interestingly, we found that the expression of ORC1L and ORC6L in BC is consistent with levels observed in lung adenocarcinoma, glioma and gastrointestinal malignancies, such as gastric and colorectal cancer.12,15,17,21 We also found that both ORC1L and ORC6L are useful diagnostic indicators of BC. Our results suggest that ORC1L and ORC6L are potential oncogenes and may be clinically useful in BC diagnosis.
Our Kaplan–Meier analysis demonstrated that ORC expression levels were not associated with OS in BC patients, which was inconsistent with Kaplan–Meier Plotter’s prediction. However, using a Cox regression analysis, which accounted for potentially confounding clinicopathological factors including age and molecular subtype of BC, we confirmed that ORC6L is an independent prognostic factor in this disease. ORC1L and ORC5L had no significant effect on OS, despite their differential expression in BC versus healthy tissue.
Evidence suggests that ORC6L is a potential oncogenic gene in BC and could be used as a potential prognostic factor or novel therapeutic target to improve survival in people with BC. However, the association of ORC5L expression with BC is unclear and its diagnostic value is relatively low. More evidence is needed to confirm whether and how this gene is differentially expressed in BC. The ORC2L and ORC3L proteins were abnormally expressed in BC, but there were no significant differences in their corresponding mRNA levels, and their expression had no effect OS in this study. ORC4L was not differentially expressed at either the protein or the mRNA level. In light of the significant differential expression and clinical significance of ORC6L, we constructed a nomogram to predict the probability of survival in people with BC at 1, 3, and 5 years, which was found to be highly accurate and of high clinical value.
Transcriptional profile comparisons of high versus low ORC6L expression using GSEA led us to identify a range of pathways (including those involved in cell cycle checkpoints and cellular senescence) that were associated with the expression of this gene. Other pathways, including such as NF-κB, P53, and WNT, were also activated with higher ORC6L expression. The effects of these pathways are to promote proliferation, angiogenesis, stem cell characteristics, and tumor migration. However, the exact molecular mechanism by which ORC6L acts in BC requires validation in clinical BC specimens and animal models in future to confirm these findings.
Cumulative evidence suggests that tumor-infiltrating immune cell plays essential roles in BC development and progression.31 Th1 and Th2 cytokines have been shown to contribute to tumorigenesis in several models. Functional Th1-oriented T follicular helper tumor-infiltrating lymphocytes, which infiltrate human BC, promote effective adaptive immunity.32 Th2 activation is associated with poor prognosis in BC.33 Recently, Ghirelli et al described a novel granulocyte-macrophage colony-stimulating factor (GM-CSF)/pDC/Th2 axis in aggressive subtypes of BC.34 Treg cells, which are critical for anti-tumor immunity, play a major role in the development of an immunosuppressive tumor microenvironment. Studies have shown that increased numbers of tumor-infiltrating Treg cells correlate with reduced survival, and quantification of Tregs therefore enables the identification of those with high-risk BC and those at risk of late relapse.35 We found that the high levels of ORC6L expression were associated with increased Th1, Th2 and Treg cell infiltration, indicating that ORC6L might improve the ratio of the immune cells that infiltrate the tumor in BC through an as yet unknown mechanism, in favour of reducing tumor progression and improving clinical prognosis.
The evidence for pro- and antitumor effects of mast cells in BC progression remains conflicting.36 Studies have showed that mast cells increase BC growth and metastasis,37 but a study by della Rovere et al has revealed that peritumoral mast cells have a cytolytic activity against tumor cells.38 Moreover, decreased NK-cell tumor immunosurveillance was found to enhance metastasis in BC models.39 We found that infiltration of mast cells and NK cells was reduced in BC tissues with high ORC6L expression, suggesting that inhibition of tumor infiltration by ORC6L is another factor that may promote BC progression. Further studies are needed to evaluate the role of these immune cells within the BC microenvironment.
Conclusions
We demonstrated for the first time that ORC1L, ORC5L and ORC6L are differentially expressed in BC and have diagnostic value. Among them, ORC6L has the highest diagnostic value and is an independent risk factor that affects OS in people with BC, and could thus be used as a potential prognostic biomarker. As the expression, and diagnostic and prognostic values of ORC subunits in BC in this study were based on data from public databases, in vitro and in vivo experiments and clinical studies are required to confirm our findings.
Abbreviations
AUC, area under the curve; BC, breast cancer; CI, confidence interval; HR, hazard ratio; ORC, origin recognition complex; OS, overall survival; ROC, receiver operating characteristic.
Data Sharing Statement
The raw data used in this study were derived from TCGA (https://portal.gdc.cancer.gov/), GEO (https://www.ncbi.nlm.nih.gov/geo/), Oncomine (https://www.oncomine.org/), UALCAN (http://ualcan.path.uab.edu/) and Kaplan–Meier plotter (http://kmplot.com/analysis/) databases, which are publicly available.
Ethics Approval and Informed Consent
All data for this study were obtained from the TCGA, GEO, Oncomine, UALCAN and Kaplan–Meier plotter databases, which are publicly available. We did not obtain these data directly from patients or animals. Therefore, following ethical review, the Affiliated Hospital of Guilin Medical University Ethics Committee granted a status of exemption.
Consent for Publication
All authors gave final approval to submit the manuscript for publication.
Acknowledgments
We thank those involved in the compilation and maintenance of the databases used in this study, which made our analyses possible.
Funding
This research was supported by the National Natural Science Foundation of China (No. 81802478), the Guangxi Natural Science Foundation (No. 2018GXNSFBA050042) and the Guangxi Science and Technology Base and Talents special project (No. AD20238005).
Disclosure
The authors declare that they have no competing interests.
References
1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
2. Cardoso F, Fallowfield L, Costa A, Castiglione M, Senkus E. Locally recurrent or metastatic breast cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2011;22(Suppl 6):vi25–vi30. doi:10.1093/annonc/mdr372
3. Suo HD, Tao Z, Zhang L, et al. Coexpression network analysis of genes related to the characteristics of tumor stemness in triple-negative breast cancer. BioMed Res Int. 2020;2020:7575862. doi:10.1155/2020/7575862
4. Koleck TA, Conley YP. Identification and prioritization of candidate genes for symptom variability in breast cancer survivors based on disease characteristics at the cellular level. Breast Cancer. 2016;8:29–37. doi:10.2147/BCTT.S88434
5. Bell SP, Dutta A. DNA replication in eukaryotic cells. Ann Rev Biochem. 2002;71:333–374. doi:10.1146/annurev.biochem.71.110601.135425
6. Prasanth SG, Shen Z, Prasanth KV, Stillman B. Human origin recognition complex is essential for HP1 binding to chromatin and heterochromatin organization. Proc Natl Acad Sci. 2010;107(34):15093–15098. doi:10.1073/pnas.1009945107
7. Prasanth SG, Prasanth KV, Stillman B. Orc6 involved in DNA replication, chromosome segregation, and cytokinesis. Science. 2002;297(5583):1026–1031. doi:10.1126/science.1072802
8. Dhar SK, Yoshida K, Machida Y, et al. Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell. 2001;106(3):287–296. doi:10.1016/S0092-8674(01)00458-5
9. Kordon MM, Szczurek A, Berniak K, et al. PML-like subnuclear bodies, containing XRCC1, juxtaposed to DNA replication-based single-strand breaks. FASEB J. 2019;33(2):2301–2313. doi:10.1096/fj.201801379R
10. Koleck TA, Bender CM, Clark BZ, et al. An exploratory study of host polymorphisms in genes that clinically characterize breast cancer tumors and pretreatment cognitive performance in breast cancer survivors. Breast Cancer. 2017;9:95–110. doi:10.2147/BCTT.S123785
11. El-Saghire H, Vandevoorde C, Ost P, et al. Intensity modulated radiotherapy induces pro-inflammatory and pro-survival responses in prostate cancer patients. Int J Oncol. 2014;44(4):1073–1083. doi:10.3892/ijo.2014.2260
12. Xiong W, Xie C, Qiu Y, Tu Z, Gong Q. Origin recognition complex subunit 1 regulates cell growth and metastasis in glioma by altering activation of ERK and JNK signaling pathway. Mol Cell Probes. 2020;49:101496. doi:10.1016/j.mcp.2019.101496
13. Liu Y, Chen P, Li M, et al. Comprehensive analysis of the control of cancer stem cell characteristics in endometrial cancer by network analysis. Comput Math Methods Med. 2021;2021:6653295. doi:10.1155/2021/6653295
14. Gavin EJ, Song B, Wang Y, Xi Y, Ju J. Reduction of Orc6 expression sensitizes human colon cancer cells to 5-fluorouracil and cisplatin. PLoS One. 2008;3(12):e4054. doi:10.1371/journal.pone.0004054
15. Xi Y, Formentini A, Nakajima G, Kornmann M, Ju J. Validation of biomarkers associated with 5-fluorouracil and thymidylate synthase in colorectal cancer. Oncol Rep. 2008;19(1):257–262.
16. Hu Y, Wang L, Li Z, et al. Potential prognostic and diagnostic values of CDC6, CDC45, ORC6 and SNHG7 in colorectal cancer. Onco Targets Ther. 2019;12:11609–11621. doi:10.2147/OTT.S231941
17. Mao R, Wang Z, Zhang Y, et al. Development and validation of a novel prognostic signature in gastric adenocarcinoma. Aging. 2020;12(21):22233–22252. doi:10.18632/aging.104161
18. Zhou W, Li J, Lu X, et al. Derivation and validation of a prognostic model for cancer dependency genes based on CRISPR-Cas9 in gastric adenocarcinoma. Fron Oncol. 2021;11:617289. doi:10.3389/fonc.2021.617289
19. Wang XK, Wang QQ, Huang JL, et al. Novel candidate biomarkers of origin recognition complex 1, 5 and 6 for survival surveillance in patients with hepatocellular carcinoma. J Cancer. 2020;11(7):1869–1882. doi:10.7150/jca.39163
20. Nangraj AS, Selvaraj G, Kaliamurthi S, Kaushik AC, Cho WC, Wei DQ. Integrated PPI- and WGCNA-retrieval of hub gene signatures shared between Barrett’s esophagus and esophageal adenocarcinoma. Front Pharmacol. 2020;11:881. doi:10.3389/fphar.2020.00881
21. Zhang Y, Tseng JT, Lien IC, Li F, Wu W, Li H. mRNAsi index: machine learning in mining lung adenocarcinoma stem cell biomarkers. Genes. 2020;11(3):257.
22. Song B, Liu XS, Rice SJ, et al. Plk1 phosphorylation of orc2 and hbo1 contributes to gemcitabine resistance in pancreatic cancer. Mol Cancer Ther. 2013;12(1):58–68. doi:10.1158/1535-7163.MCT-12-0632
23. Kim SH, Ho JN, Jin H, et al. Upregulated expression of BCL2, MCM7, and CCNE1 indicate cisplatin-resistance in the set of two human bladder cancer cell lines: T24 cisplatin sensitive and T24R2 cisplatin resistant bladder cancer cell lines. Invest Clin Urol. 2016;57(1):63–72. doi:10.4111/icu.2016.57.1.63
24. Ősz Á, Lánczky A, Győrffy B. Survival analysis in breast cancer using proteomic data from four independent datasets. Sci Rep. 2021;11(1):16787. doi:10.1038/s41598-021-96340-5
25. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi:10.1016/j.neo.2017.05.002
26. Liu J, Lichtenberg T, Hoadley KA, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173(2):400–416.e411. doi:10.1016/j.cell.2018.02.052
27. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi:10.1073/pnas.0506580102
28. Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. doi:10.1186/1471-2105-14-7
29. Deepak KGK, Vempati R, Nagaraju GP, et al. Tumor microenvironment: challenges and opportunities in targeting metastasis of triple negative breast cancer. Pharmacol Res. 2020;153:104683. doi:10.1016/j.phrs.2020.104683
30. Mittal S, Brown NJ, Holen I. The breast tumor microenvironment: role in cancer development, progression and response to therapy. Expert Rev Mol Diagn. 2018;18(3):227–243. doi:10.1080/14737159.2018.1439382
31. Stanton SE, Disis ML. Clinical significance of tumor-infiltrating lymphocytes in breast cancer. J Immunother Cancer. 2016;4:59. doi:10.1186/s40425-016-0165-6
32. Noël G, Langouo Fontsa M, Garaud S, et al. Functional Th1-oriented T follicular helper cells that infiltrate human breast cancer promote effective adaptive immunity. J Clin Invest. 2021;131(19):e139905. doi:10.1172/JCI139905
33. Teschendorff AE, Gomez S, Arenas A, et al. Improved prognostic classification of breast cancer defined by antagonistic activation patterns of immune response pathway modules. BMC Cancer. 2010;10:604. doi:10.1186/1471-2407-10-604
34. Ghirelli C, Reyal F, Jeanmougin M, et al. Breast cancer cell-derived GM-CSF licenses regulatory Th2 induction by plasmacytoid predendritic cells in aggressive disease subtypes. Cancer Res. 2015;75(14):2775–2787. doi:10.1158/0008-5472.CAN-14-2386
35. Bates GJ, Fox SB, Han C, et al. Quantification of regulatory T cells enables the identification of high-risk breast cancer patients and those at risk of late relapse. J Clin Oncol. 2006;24(34):5373–5380. doi:10.1200/JCO.2006.05.9584
36. Aponte-López A, Fuentes-Pananá EM, Cortes-Muñoz D, Muñoz-Cruz S. Mast cell, the neglected member of the tumor microenvironment: role in breast cancer. J Immunol Res. 2018;2018:2584243. doi:10.1155/2018/2584243
37. Majorini MT, Cancila V, Rigoni A, et al. Infiltrating mast cell-mediated stimulation of estrogen receptor activity in breast cancer cells promotes the luminal phenotype. Cancer Res. 2020;80(11):2311–2324. doi:10.1158/0008-5472.CAN-19-3596
38. Della Rovere F, Granata A, Familiari D, D’Arrigo G, Mondello B, Basile G. Mast cells in invasive ductal breast cancer: different behavior in high and minimum hormone-receptive cancers. Anticancer Res. 2007;27(4b):2465–2471.
39. Bottos A, Gotthardt D, Gill JW, et al. Decreased NK-cell tumour immunosurveillance consequent to JAK inhibition enhances metastasis in breast cancer models. Nat Commun. 2016;7:12258. doi:10.1038/ncomms12258
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.