Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 16
Exploration into Plasma Hsa_circ_0052184 as a New Biomarker of Colorectal Cancer Prognosis
Received 22 March 2023
Accepted for publication 26 May 2023
Published 12 June 2023 Volume 2023:16 Pages 589—597
DOI https://doi.org/10.2147/PGPM.S413451
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin H Bluth
Enqi Zheng, Deshuang Xiao
Hernia Vascular Surgery, The First People’s Hospital of Wenling, Wenling, Zhejiang, People’s Republic of China
Correspondence: Deshuang Xiao, Hernia Vascular Surgery, The First People’s Hospital of Wenling, No. 333 Chuan’an South Road, Chengxi Street, Wenling, 317599, Zhejiang, People’s Republic of China, Tel +86-057689668595, Email [email protected]
Background: Circular RNAs (circRNAs) are strong modulators of tumor pathology. Herein, our goal was to examine the plasma hsa_circ_0052184 content among colorectal cancer (CRC) patients, and assess its association with patient clinicopathological profile and diagnostic values.
Methods: Overall, we collected 228 presurgical CRC and 146 normal plasma samples from The First People’s Hospital of Wenling. Circulating hsa_circ_0052184 levels were assessed via qRT-PCR, and the diagnostic prediction was conducted with the receiver operating characteristic (ROC) curve.
Results: Relative to healthy controls, CRC patients exhibited markedly enhanced circulating hsa_circ_0052184 levels, which were closely correlated with advanced stage of disease and worse outcome. Based on our uni- (UA) and multivariate assessments (MA), elevated hsa_circ_0052184 levels were a stand-alone predictor of poor prognosis. The ROC curve depicted an area under the curve (AUC) for CRC diagnosis to be 0.9072.
Conclusion: Circulating hsa_circ_0052184 is a potential bioindicator of CRC outcome.
Keywords: hsa_circ_0052184, diagnostic biomarker, colorectal cancer
Introduction
Colorectal cancer (CRC) is the third leading form of global malignancy, and is the second contributor to cancer-associated deaths.1 Based on reports from the Global Cancer Statistics 2020, the year 2020 alone brought over 1.9 million new CRC incidences, with 935,000 associated fatalities from around the world. This accounted for approximately 10% of all cancer cases and associated mortalities.2 Common CRC therapies include surgery, accompanied by radio-, chemo-, or targeted therapies. Notably, older patients with T4 CRC were more prone to severe postoperative complications, but age did not impact survival outcomes. For this reason, older patients should not be denied surgery for T4 CRC based on age alone.3 Employing endogenous biomarkers in cancer diagnosis, like in CRC, holds great significance.4,5 More recently, liquid biopsy is increasingly employed as a non-invasive approach to disease diagnosis in hospitals.6 Moreover, with advancements in high-throughput omics (for example, genomics, proteomics, and metabolomics) as well as development of novel identification protocols, scientists have uncovered several novel tumor biomarkers, particularly, those involving noncoding RNAs (ncRNAs).7–9 In contrast, linear RNAs are relatively unstable in vitro, and degrade easily, which restricts its clinical usage.
Circular RNAs (circRNAs) are a newly discovered group of ncRNAs, with a characteristic stable and circular configuration.10–12 Several reports suggest circRNAs as potential disease bioindicators owing to their tissue- and disease stage-specific expression profile.13,14 Their primary action is gene regulation via modulation of micro RNAs (miRNAs)15 and protein.14 Owing to their contribution to several cell transduction networks, circRNAs may be employed as diagnostic and/or prognostic agents in multiple cancers, including CRC.16,17 Among its unique characteristics is its covalent cyclic configuration, which allows it to escape digestion by exonucleases. Moreover, its relatively stable nature, especially in body fluids (namely, blood, urine, and saliva) it allows it to be a superior disease biomarker, compared to linear RNAs.18
Human hsa_circ_0052184 is a newly discovered circRNA harboring 221bp nt in spliced sequence length. Its gene resides on chr19:55603809–55604030, and its gene symbol is PPP1R12C. Herein, we attempted to assess the diagnostic power, as well as the tissue and plasma sensitivity of hsa_circ_0052184 in early-stage CRC diagnosis.
Materials and Methods
Samples Accumulation
Overall, we collected 228 presurgical CRC and 146 normal plasma samples from The First People’s Hospital of Wenling. The following patients were included in our analysis: those (a) with primary CRC diagnosis, as evidenced by pathological examination; (b) between 18–80 years of age; (c) with complete tumor and survival information; and (d) with no neoadjuvant treatment prior to operation. Upon collection, samples were maintained at −80°C until further analysis. The research strategy was formulated in accordance with the Declaration of Helsinki. This research received ethical approval (KY-2023-1004-01) from the The First People’s Hospital of Wenling, and received informed consent from all participants before initiation of the investigation.
Quantitative Real-Time Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
Total RNA isolation was conducted from collected plasma samples via TRIzol (Invitrogen), following kit protocols. Following RNA quantification and quality check via NanoDrop ND2000 spectrophotometer (NanoDrop), 1 µg total RNA (in 20 µL mixture) was converted to cDNA via PrimeScript RT-polymerase (Takara), followed by qRT-PCR with SYBR Green Premix Ex Taq (Takara Bio) and the ABI PRISM 7500 Sequence Detection System (Applied Biosystems, Life Technologies). Relative circRNAs levels were computed subsequently.
Cell Culture
The CRC (HCT116, HT29, SW480, and SW620) and normal human colonic epithelial cells (NCM460) were purchased from Biobw (Beijing, China) and maintained in DMEM (Dulbecco’s modified Eagle medium) with 10% FBS (fetal bovine serum) (Gibco, NY, USA) in an incubator with 5% CO2 and at 37°C.
Cell Counting Kit-8 (CCK-8) Assay
Cell survival was assessed via the CCK-8 assay (Beyotime; C0037), following kit directions. In brief, 1000 cells/well were seeded in 96-well plates in medium containing 10% FBS and penicillin-streptomycin (PS) at 37 °C with 5% CO2. Absorbance was recorded at 450nm (reference wavelength: 650nm) via a microplate reader (Bio-Rad).
Statistical Analysis
GraphPad 7.0 (GraphPad, Inc., CA, USA) and SPSS 16.0 software, in particular, the Chi-square and Student’s t-tests were employed for data analyses, as needed. Pearson’s correlation analyses were utilized for assessing variable-to-variable associations. Hsa_circ_0052184 AUC values, sensitivity, and specificity were computed via the receiver operating characteristic (ROC) analysis, which provided the predictive performance of hsa_circ_0052184 in delineating between CRC patients and healthy volunteers. p < 0.05 was regarded as significant.
Results
Hsa_circ_0052184 Was Highly Expressed Among CRC Plasma Samples
Our analysis of the GEO dataset (GSE172229) revealed that the circRNAs accurately differentiated between CRC samples and paracancerous tissues (Figure 1A). More importantly, hsa_circ_0052184 displayed the highest level of expression among CRC versus normal tissue levels (Figure 1B). Hence, hsa_circ_0052184 was chosen for subsequent analysis. As expected, hsa_circ_0052184 was highly expressed among CRC tissues, as opposed to adjacent healthy tissues (Figure 1C). Moreover, hsa_circ_0052184 was strongly upregulated in all CRC versus normal colon epithelial cells (NCM460, Figure 1D). Additionally, the circulating hsa_circ_0052184 levels were markedly enhanced among CRC patients, as opposed to healthy controls (Figure 1E). Herein, we demonstrated a direct relationship between the hsa_circ_0052184 content in CRC tissues and its levels in the circulation (Figure 1F). Hence, hsa_circ_0052184 may potentially be released into circulation by tumors.
Baseline Profiles and Hsa_circ_0052184 Content in 228 CRC Patients
We also assessed the circulating hsa_circ_0052184 content in a validation cohort involving 228 CRC patients. Using correlation analysis, we demonstrated a strong association between plasma hsa_circ_0052184 levels and pN status, pM status, clinical stage and tumor size. Of note, there was no correlation between plasma hsa_circ_0052184 contents and other patient variables like age, gender, tumor location and pT status (Table 1). Moreover, circulating hsa_circ_0052184 contents were strongly correlated with diminished progression-free survival (PFS, Figure 2A) and overall survival (OS, Figure 2B) among CRC patients with elevated versus reduced hsa_circ_0052184 expression. Similarly, plasma hsa_circ_0052184 content was associated with PFS (Table 2) and OS (Table 3) in univariate analyses (UA), as well as strongly diminished patient PFS (Table 2) and OS (Table 3) in multivariate analyses (MA).
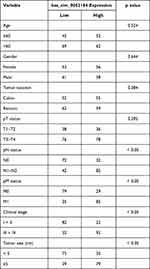 |
Table 1 Association Between Hsa_circ_0052184 Expression and Clinicopathological Features |
 |
Table 2 Uni- (UA) and Multivariate Cox Analyses (MA) of Factors Related to the CRC Patient PFS |
 |
Table 3 Uni- (UA) and Multivariate Cox Analyses (MA) of Factors Related to the CRC Patient OS |
 |
Figure 2 Baseline profile and hsa_circ_0052184 content of 228 CRC patients. Elevated plasma hsa_circ_0052184 levels were linked to significantly reduced PFS (A) and OS (B) among CRC patients. |
ROC Curve Assessment of the Predictive Performance of Hsa_circ_0052184
To assess the diagnosability of hsa_circ_0052184 in the early-stage CRC diagnosis, we employed ROC curve analysis. Based on our observation, The hsa_circ_0052184 AUC was 0.9072 (95% CI 0.8769–0.9374) among CRC patients, thus indicating that it may be a robust potential indicator of CRC early diagnosis (Figure 3).
Hsa_circ_0052184 Deficiency Inhibited CRC Cell Proliferation and Metastasis
To elucidate the underlying mechanism behind hsa_circ_0052184 action in CRC, we silenced hsa_circ_0052184 in the HCT116 and SW620 cells (Figure 4A). Based on our CCK-8 analysis, hsa_circ_0052184 deficiency strongly suppressed cell proliferation (Figure 4B). Moreover, using transwell assay, we demonstrated that hsa_circ_0052184 silencing strongly inhibited HCT116 and SW620 cell migration and invasion (Figure 4C and D). Taken together, we revealed that hsa_circ_0052184 knockdown substantially reduces breast cancer cell proliferation and metastasis.
Discussion
CRC is a widespread cancer with an elevated mortality rate, and it is mostly identified at a progressed stage of disease.19 Microsatellite instability (MSI) is the molecular fingerprint of a deficient mismatch repair system. Approximately 15% of CRC display MSI owing either to epigenetic silencing of MLH1 or a germline mutation in one of the mismatch repair genes MLH1, MSH2, MSH6 or PMS2.20 Moreover, TP53 mutation has been found in about 43% of sporadic CRC cases (IARC TP53 database; https://p53.iarc.fr), and MSI-L CRC patients show higher incidence of KRAS mutations.21 Thus, screening for defective, DNA mismatch repair in CRC patients should include immunohistochemistry (IHC) and/or MSI test.22,23 However, Serum biomarker identification is among the most ambitious pursuits of the current oncologic research.24 At present, there are a few available biomarkers for CRC detection. However, these are not highly sensitive or specific, such as circulating tumor cells, cfDNA, etc.25 Hence, over the last 3–4 years,26 there has been much research on the application of circRNAs as potential bioindicators for certain tumors like breast cancer27 and CRC,16 primarily due to their enhanced stability in bodily fluids.28
CircRNAs are a novel group of O-shaped RNAs available within living cells. Compared to the classical linear RNAs, circRNAs do not undergo exonuclease- and RNase-mediated degradation as they lack the 5′ end, 3′ end, and poly(A) tail.29 Owing to this unique feature, circRNAs have long half-lives, and can therefore be effective biomarker candidates. Moreover, human circRNA molecules are present in 10 times larger quantities than homogenetic linear isomer RNA molecules.30 CircRNAs contain highly conserved sequences, long half-life, and tissue-specificity. Additionally, they are known to post-transcriptionally modulate gene expression5,31 by sequestering target mRNAs.32 Till now, multiple differentially regulated circRNAs have been identified in various tissues, blood,33 saliva,34 and other bodily fluid,35 thereby indicating their candidacies as bioindicators in several diseases. CircRNAs, in combination other reported biomarkers, may enhance the accuracy of certain disease diagnoses. However, a majority of these studies investigated potential functions, while the circRNA-based diagnostic performance remains largely undetermined in CRC.
Herein, we demonstrated elevated hsa_circ_0052184 levels in CRC plasma and tissues, as opposed to controls. Moreover, we showed a direct association between the high hsa_circ_0052184 content between tissues and circulation. This suggests the possibility of tumor cells releasing hsa_circ_0052184 into the circulation. Subsequently, using Kaplan–Meier analysis, we revealed that the elevated hsa_circ_0052184 content among CRC patients was intricately linked to worse outcome. Additionally, our UA and MA revealed that elevated hsa_circ_0052184 content was a stand-alone indicator of CRC risk. Hsa_circ_0052184 originates from the noncoding region of PPP1R12C, and it spans 221bp. Our findings revealed a strong tumorigenic role of hsa_circ_0052184 in CRC progression. Based on our ROC curves, circulating hsa_circ_0052184 has superior predictive performance for CRC diagnosis, thereby indicating that hsa_circ_0052184 has great potential as a robust and widely applied tumor biomarker. However, the limitation is that we have not conducted further research on the mechanism of hsa_circ_0052184 in the progression of CRC.
Conclusion
In conclusion, herein, we revealed that circulating hsa_circ_0052184 content is relatively high among CRC patients, and it has potent predictive performance in early CRC diagnosis. Hence, circulating hsa_circ_0052184 can be employed as a potential candidate for CRC diagnosis.
Funding
There is no funding to report.
Disclosure
No conflicts of interest to declare.
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. doi:10.3322/caac.21654
2. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
3. Osseis M, Nehmeh WA, Rassy N, et al. Surgery for T4 colorectal cancer in older patients: determinants of outcomes. J Pers Med. 2022;12(9):9. doi:10.3390/jpm12091534
4. Kim EJ, Baik GH. 만성위염, 장상피화생, 위암 환자의 위점막 상재균 변화의 차이에 대한 고찰 [Review on gastric mucosal microbiota profiling differences in patients with chronic gastritis, intestinal metaplasia, and gastric cancer]. Korean J Gastroenterol. 2014;64(6):390–393. Korean. doi:10.4166/kjg.2014.64.6.390
5. Shin VY, Chu KM. MiRNA as potential biomarkers and therapeutic targets for gastric cancer. World J Gastroenterol. 2014;20(30):10432–10439. doi:10.3748/wjg.v20.i30.10432
6. Liu X, Cai H, Wang Y. Prognostic significance of tumour markers in Chinese patients with gastric cancer. ANZ J Surg. 2014;84(6):448–453. doi:10.1111/j.1445-2197.2012.06287.x
7. Takayasu T, Shah M, Dono A, et al. Cerebrospinal fluid ctDNA and metabolites are informative biomarkers for the evaluation of CNS germ cell tumors. Sci Rep. 2020;10(1):14326. doi:10.1038/s41598-020-71161-0
8. Guo LY, Wu AH, Wang YX, Zhang LP, Chai H, Liang XF. Deep learning-based ovarian cancer subtypes identification using multi-omics data. BioData Min. 2020;13:10. doi:10.1186/s13040-020-00222-x
9. Rahat B, Ali T, Sapehia D, Mahajan A, Kaur J. Circulating cell-free nucleic acids as epigenetic biomarkers in precision medicine. Front Genet. 2020;11:844. doi:10.3389/fgene.2020.00844
10. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi:10.1261/rna.035667.112
11. Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143(11):1838–1847. doi:10.1242/dev.128074
12. Memczak S, Papavasileiou P, Peters O, Rajewsky N, Pfeffer S. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One. 2015;10(10):e0141214. doi:10.1371/journal.pone.0141214
13. Fan L, Li W, Jiang H. Circ_0000395 promoted CRC progression via elevating MYH9 expression by sequestering miR-432-5p. Biochem Genet. 2022;61(1):116–137. doi:10.1007/s10528-022-10245-0
14. Wang X, Chen M, Fang L. hsa_circ_0068631 promotes breast cancer progression through c-Myc by binding to EIF4A3. Mol Ther Nucleic Acids. 2021;26:122–134. doi:10.1016/j.omtn.2021.07.003
15. Bi J, Liu H, Cai Z, et al. Circ-BPTF promotes bladder cancer progression and recurrence through the miR-31-5p/RAB27A axis. Aging. 2018;10(8):1964–1976. doi:10.18632/aging.101520
16. Yang H, Li X, Meng Q, et al. CircPTK2 (hsa_circ_0005273) as a novel therapeutic target for metastatic colorectal cancer. Mol Cancer. 2020;19(1):13. doi:10.1186/s12943-020-1139-3
17. Tian J, Xi X, Wang J, et al. CircRNA hsa_circ_0004585 as a potential biomarker for colorectal cancer. Cancer Manag Res. 2019;11:5413–5423. doi:10.2147/CMAR.S199436
18. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi:10.1038/nature11928
19. Moreno CC, Mittal PK, Sullivan PS, et al. Colorectal cancer initial diagnosis: screening colonoscopy, diagnostic colonoscopy, or emergent surgery, and tumor stage and size at initial presentation. Clin Colorectal Cancer. 2016;15(1):67–73. doi:10.1016/j.clcc.2015.07.004
20. Vilar E, Gruber SB. Microsatellite instability in colorectal cancer-The stable evidence. Nat Rev Clin Oncol. 2010;7(3):153–162. doi:10.1038/nrclinonc.2009.237
21. Oliart S, Martinez-Santos C, Moreno-Azcoita M, et al. Do MSI-L sporadic colorectal tumors develop through “mild mutator pathway”? Am J Clin Oncol. 2006;29(4):364–370. doi:10.1097/01.coc.0000221428.35366.cb
22. Adeleke S, Haslam A, Choy A, et al. Microsatellite instability testing in colorectal patients with Lynch syndrome: lessons learned from a case report and how to avoid such pitfalls. Per Med. 2022;19(4):277–286. doi:10.2217/pme-2021-0128
23. Mathews NS, Masih D, Mittal R, et al. Microsatellite instability in young patients with mucinous colorectal cancers - characterization using molecular testing, immunohistochemistry, and histological features. Indian J Cancer. 2019;56(4):309–314. doi:10.4103/ijc.IJC_224_18
24. Pastore AL, Palleschi G, Silvestri L, et al. Serum and urine biomarkers for human renal cell carcinoma. Dis Markers. 2015;2015:251403. doi:10.1155/2015/251403
25. Boussios S, Ozturk MA, Moschetta M, et al. The developing story of predictive biomarkers in colorectal cancer. J Pers Med. 2019;9(1):1. doi:10.3390/jpm9010012
26. Piawah S, Venook AP. Targeted therapy for colorectal cancer metastases: a review of current methods of molecularly targeted therapy and the use of tumor biomarkers in the treatment of metastatic colorectal cancer. Cancer. 2019;125(23):4139–4147. doi:10.1002/cncr.32163
27. Omid-Shafaat R, Moayeri H, Rahimi K, et al. Serum Circ-FAF1/Circ-ELP3: a novel potential biomarker for breast cancer diagnosis. J Clin Lab Anal. 2021;35(11):e24008. doi:10.1002/jcla.24008
28. Chen Y, Li C, Tan C, Liu X. Circular RNAs: a new frontier in the study of human diseases. J Med Genet. 2016;53(6):359–365. doi:10.1136/jmedgenet-2016-103758
29. Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148. doi:10.1016/j.canlet.2015.06.003
30. Yu CY, Kuo HC. The emerging roles and functions of circular RNAs and their generation. J Biomed Sci. 2019;26(1):29. doi:10.1186/s12929-019-0523-z
31. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
32. Chen LY, Wang L, Ren YX, et al. The circular RNA circ-ERBIN promotes growth and metastasis of colorectal cancer by miR-125a-5p and miR-138-5p/4EBP-1 mediated cap-independent HIF-1alpha translation. Mol Cancer. 2020;19(1):164. doi:10.1186/s12943-020-01272-9
33. Zhang YG, Yang HL, Long Y, Li WL. Circular RNA in blood corpuscles combined with plasma protein factor for early prediction of pre-eclampsia. BJOG. 2016;123(13):2113–2118. doi:10.1111/1471-0528.13897
34. Bahn JH, Zhang Q, Li F, et al. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem. 2015;61(1):221–230. doi:10.1373/clinchem.2014.230433
35. Qu S, Zhong Y, Shang R, et al. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14(8):992–999. doi:10.1080/15476286.2016.1220473
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



