Back to Journals » Journal of Asthma and Allergy » Volume 16
Exploration and Validation of Potential Biomarkers and Therapeutic Targets in Ferroptosis of Asthma
Authors Xing Y , Feng L , Dong Y, Li Y, Zhang L, Wu Q, Huo R, Dong Y, Tian X , Tian X
Received 7 April 2023
Accepted for publication 6 July 2023
Published 12 July 2023 Volume 2023:16 Pages 689—710
DOI https://doi.org/10.2147/JAA.S416276
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Luis Garcia-Marcos
Yanqing Xing,1 Liting Feng,1 Yangdou Dong,2 Yupeng Li,1 Lulu Zhang,1 Qiannan Wu,1 Rujie Huo,1 Yanting Dong,1 Xinrui Tian,1 Xinli Tian3
1Department of Respiratory and Critical Care Medicine, The Second Hospital of Shanxi Medical University, Taiyuan, People’s Republic of China; 2College of Basic Medicine, Shanxi Medical University, Taiyuan, People’s Republic of China; 3Department of Cardiology, Chinese PLA General Hospital, Beijing, People’s Republic of China
Correspondence: Xinrui Tian, Department of Respiratory and Critical Care Medicine, The Second Hospital of Shanxi Medical University, No. 382, Wuyi Road, Xinghualing District, Taiyuan, People’s Republic of China, Tel +8613834575570, Email [email protected] Xinli Tian, Department of Cardiology, Chinese PLA General Hospital, No. 7 Medical Center No. 5 Nanmencang, Dongsijiao, Dongcheng District, Beijing, People’s Republic of China, Email [email protected]
Purpose: Asthma is a chronic inflammatory airway disease involving multiple mechanisms, of which ferroptosis is a form of programmed cell death. Recent studies have shown that ferroptosis may play a crucial role in the pathogenesis of asthma, but no specific ferroptosis gene has been found in asthma, and the exact mechanism is still unclear. The present study aimed to screen ferroptosis genes associated with asthma and find therapeutic targets, in order to contribute a new clue for the diagnosis and therapy of asthma.
Methods: Ferroptosis-related differentially expressed genes (FR-DEGs) in asthma were selected by the GSE41861, GSE43696 and ferroptosis datasets. Next, the FR-DEGs were subjected by GO and KEGG enrichment, and the mRNA-miRNA network was constructed. Then, GSEA and GSVA enrichment analysis and Immune infiltration analysis were performed, followed by targeted drug prediction. Finally, the expression of FR-DEGs was confirmed using GSE63142 dataset and RT-PCR assay.
Results: We found 13 FR-DEGs by the GSE41861, GSE43696 and ferroptosis database. Functional enrichment analysis revealed that the 13 FR-DEGs were enriched in oxidative stress, immune response, ferroptosis, lysosome, necrosis, apoptosis etc. Moreover, our results revealed the mRNA-miRNA network of the FR-DEGs and identified candidate drugs. Also, immune infiltration revealed that ELAVL1, CREB5, CBR1 and NR1D2 are associated with the immune cells and may be potential targets in asthma. Finally, 10 FR-DEGs were validated by the GSE63142 database. It was verified that 7 FR-DEGs were differentially expressed by collecting asthma patients and healthy controls.
Conclusion: This study ultimately identified 7 FR-DEGs for the diagnosis and therapy of asthma. These 7 FR-DEGs contribute to oxidative stress and immune responses. This study provides potential therapeutic targets and biomarkers for asthma patients, shedding further light on the pathogenesis of asthma as well as providing new insights into the treatment of asthma.
Keywords: asthma, bioinformatics, ferroptosis, GSEA, GSVA, immune microenvironment
Introduction
Asthma is an immune-related chronic airway inflammatory disease with major pathological changes including airway inflammation, smooth muscle dysfunction and airway remodeling with progressively increased morbidity and mortality in recent years.1 According to the 2022 Global Initiative for Asthma (GINA) guidelines,2 the common clinical phenotypes of asthma are allergic asthma, non-allergic asthma, adult-onset (late-onset) asthma, asthma with persistent airflow limitation, and asthma with obesity, and treatment guided by phenotype is clinically important, but no specific pathological features between clinical phenotype and response to treatment have been identified. The etiology of asthma is intricate, while current treatments work well to control symptoms in many patients, a large number of patients continue to suffer from worsening or treatment-related side effects.3 Therefore, it is critical to identify potential therapeutic targets for asthma, so as to offer new directions for the diagnosis, treatment and prognosis of asthma.
Ferroptosis is an iron-dependent and non-apoptotic form of programmed cell death that is symbolized by the formation of lipid peroxides from unsaturated fatty acids after catalysis by ester oxygenase or divalent iron,4 which results in cell death once excess lipid peroxides are accumulated.5 During the morphology of ferroptosis, the cell membrane and nucleus are intact, but the matrix is reduced or absent, and there is an increase in membrane density and a decrease in mitochondrial volume, which distinguishes it from other forms of cell death or apoptosis.6 The specific mechanism of ferroptosis is currently unknown and may be intricately linked to a number of biological pathways, the most well-known of which are the iron, lipid, and amino acid metabolic pathways.7–9
Ferroptosis has recently been demonstrated to be essential in many diseases.10–14 As a result, ferroptosis can be used to discover new diagnostic markers for diseases, treatments, and means of prevention, and ferroptosis has become a hot study. There are few studies on ferroptosis and asthma, and the only studies that have been conducted suggest that ferroptosis plays a very important role in the pathogenesis of asthma,15,16 but the specific mechanisms involved have not been uncovered. Some studies have shown that in the pathogenesis of asthma, reactive oxygen species (ROS) dependent lipid peroxidation and destruction of iron homeostasis can promote ferroptosis in bronchial epithelial cells, and ferrostatin-1, an ferroptosis inhibitor, has been found to reverse the process of lipid peroxidation and ferroptosis.17 It has also been found in other studies that increased levels of lipid peroxidation and ROS are accompanied by increased levels of lung ferroptosis in asthma models. Ferrostatin-1 can inhibit the ferroptosis process in asthma. These findings suggest that ferroptosis play an important role in asthma, inhibiting ferroptosis process may be a potential target for the treatment of asthma.16,18,19 In addition to ferrostatin-1, liproxstatin-1 and quercetin has also been found to alleviate asthma airway inflammation by inhibiting the ferroptosis process.20,21 Wang found that ferroptosis could be inhibited in asthma by inhibiting M1 macrophage polarization, thereby improving airway inflammation associated with ferroptosis.21 These studies suggest that ferroptosis plays an important role in the pathogenesis of asthma by regulating oxidative stress and immune response, but in the study of ferroptosis in asthma it is limited to conventional markers of ferroptosis, and no asthma-specific ferroptosis genes have been identified. Therefore, there is an urgent need to further explore the ferroptosis genes related to asthma. Further study of the relationship between asthma and ferroptosis can identify asthma-specific ferroptosis genes, which can be used as the regulator of ferroptosis in asthma and provide new therapeutic targets for asthma patients.
In view of the lack of studies on ferroptosis in asthma and the specific mechanism is unclear, we explored the ferroptosis genes related to asthma through bioinformatics to lay the foundation for the subsequent mechanism research. This is the first study to link ferroptosis-related genes to asthma, which can reflect asthma pathology. The study screened out genes that are differentially expressed in asthma by using the GSE41861 and GSE43696 databases in the Gene Expression Omnibus (GEO) database, and further extracted co- FR-DEGs as target genes. Subsequently, functional analysis of FR-DEGs in Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and Reactome pathways was performed to determine the pathogenesis of these genes in asthma. Constructed mRNA-miRNA network and predicted the target drug of the genes. Then, Gene Set Enrichment Analysis (GSEA) and Gene Set Variation Analysis (GSVA) enrichment analysis and immune landscape analysis were performed, followed by targeted drug prediction. The FR-DEGs of asthma were further validated in the microarray dataset GSE63142. Finally, differential expression levels of these ferroptosis genes were verified in asthma and healthy individuals by RT-PCR.
Materials and Methods
Data Collection and Analysis
The FerrDb (http://www.zhounan.org/ferrdb/current; last accessed August 6, 2022) database contained 565 ferroptosis-related genes (Table S1). The mRNA expression profile datasets GSE41861, GSE43696 and GSE63142 in asthma were downloaded from GEO (https://www.ncbi.nlm.nih.gov/geo/; last accessed August 12, 2022) database. A total of 138 samples (91 asthma and 47 controls) are from the GSE41861 dataset; The GSE43696 dataset contains 58 samples (38 asthma and 20 controls); GSE63142 dataset with 83 samples (56 asthma and 27 controls). FR-DEGs were screened from GSE41861, GSE43696 and FerrDb databases in asthma, and finally the FR-DEGs in asthma was verified in the GSE63142 database.
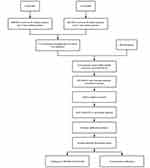
Table 1 shows the specific information of the three datasets (GSE41861, GSE43696 and GSE63142), and Figure 1 shows the flow chart of this study.
 |
Table 1 Details of GEO Asthma Date |
Differentially Expressed Genes
Microarray data and annotation files from the GSE41861 and GSE43696 datasets were downloaded, additionally, the annotation files were used to annotate the probes. Then “GEO2R” packages including “GEO query” and “R/limma” packages were used to analyze the differentially expressed genes (DEGs), and the final determined DEGs were conditioned on P value less than 0.05. The OECloud tools at https://cloud.oebiotech.com was used to create volcano maps and box line plots, whereas the “R” package was used to create Principal component analysis (PCA) plots and Venn diagrams.
From the GSE41861 and GSE43696 databases, we first extracted 272 frequently DEGs in normal and asthma samples (Table S2). Then, using the FerrDb database, we downloaded the list of ferroptosis-related genes and intersected them with 272 co-DEGs to obtain 13 ferroptosis genes between asthma and non-asthma.
GO, KEGG and Reactome Pathway Analyses of FR-DEGs
The potential functions connected to FR-DEGs were examined using the OECloud tools at https://cloud.oebiotech.com; last accessed November 14, 2022. These analyses included GO and KEGG enrichment. The species were restricted to Homo sapiens, and statistical significance was determined by an “adjusted P value less than 0.05”. These were the three criteria for the GO terms: biological process (BP), cellular component (CC), and molecular function (MF). Reactome (https://reactome.org/; last accessed October 18, 2022) is a database of biological processes and bodily responses. Finally, the results of enrichment analysis are represented by circle diagram and bubble diagram.
Construction of mRNA-miRNA Network
The ENCORI (https://starbase.sysu.edu.cn/; last accessed October 19, 2022) database was used to predict interactions between miRNAs and differentially expressed mRNAs. Then, in order to explore the potential role between miRNA and FR-DEGs’ mRNA, a miRNA-mRNA regulatory network was established. The regulatory network was visualized with the help of Cytoscape (last accessed October 19, 2022).
Single-Gene Gene Set Enrichment Analysis (GSEA)
This analysis is implemented in the GSEA package in R (last accessed October 18, 2022). In the GSEA enrichment analysis, we used the KEGG predefined signaling pathway set to detect the enrichment pathway of each FR-DEG in the GSE43696 dataset. We calculated Pearson correlation coefficients between FR-DEGs and other genes in the dataset, which was done to explore the possible signaling pathways of FR-DEGs. Table S3 contains the particular enrichment results for each FR-DEG.
Gene Set Variation Analysis (GSVA)
GSVA were performed using the OECloud tools (https://cloud.oebiotech.com; last accessed August 29, 2022). In the GSVA22 enrichment analysis, we also used the KEGG predefined signaling pathway set. To determine whether distinct metabolic pathways are enriched between samples, GSVA performed unsupervised classification of samples by converting the expression matrix of genes between samples into the expression matrix of gene sets between samples. Simultaneously, we applied the oebiotech cloud platform (https://cloud.oebiotech.com/) to analyze the difference in GSVA score of the asthma and non-asthma. Eventually p <0.05, |t|>2 is significant, in the condition of metabolic pathways were activated in asthma was t>0, pathways were restrained when t<0. The final metabolic pathway enrichment results were collected in Table S4.
Immune Infiltration Analysis
CIBERSORT is a method that uses gene expression data to characterize the abundance of cells in mixed cell populations.23 In this study, the Cibersort package (last accessed September 28, 2022) in R was used to predict the proportion of 22 types of immune infiltrating cells in the asthma immune microenvironment using the GSE41861 database. The final results are shown in Table S5.
Targeted Drug Prediction
This study used the DGIdb (https://www.dgidb.org/; last accessed November 2, 2022)24 database to predict the target drugs for FR-DEGs and to provide directions for potential drug therapy for asthma.
Validation of Ferroptosis Genes
Finally, we validated the differential expression of 13 FR-DEGs in the GSE63142 dataset and performed receiver operating characteristic (ROC) curve analysis, expressed as the area under the ROC curve (AUC). The AUC of FR-DEGs with good diagnostic efficacy was greater than 0.6.
The Expression of FR-DEGs in Asthma Patients Was Verified
Patient population: Thirty patients with asthma (meeting the criteria for asthma diagnosis) were recruited from July 2022 to January 2023 at the Second Hospital of Shanxi Medical University, and these patients were compared with thirty healthy volunteers. The Ethics Committee of the Second Hospital of Shanxi Medical University approved the study, approval number (2022) YX (079), and received informed consent for each subject. Peripheral blood was collected and peripheral blood mononuclear cells were collected by density gradient centrifugation.
RNA isolation Quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR): Total RNA from Peripheral Blood Mononuclear Cell was isolated using the RNA Isolation Kit (China, Beijing, Mei5bio MF044-01) according to the manufacturer’s protocol. RNA purity and concentration were measured using Nanodrop 2000 (ThermoFisher Scientific, USA). Next, cDNA was synthesized using the cDNA Reverse Transcription Kit (China, Nanjing, Vazyme R323-01). The specific primers were designed based on the sequence of the gene’s mRNA coding region and synthesized by Sangon Biotech. Quantitative PCR was performed using SYBR Green PCR (China, Nanjing, Vazyme Q711-02) and the details of the primers used are shown in Table 2. The relative expression of mRNA was calculated by 2−ΔΔCt method with the normalization to β-actin.
 |
Table 2 The Primers Used in This Study |
Statistical Analysis
This study was analyzed in R. The Student’s t-test was used for comparison between the asthma and controls. DEGs were analyzed using the OECloud tools at https://cloud.oebiotech.com, and the final identified DEGs were statistically significant with p-values less than 0.05. GO and KEGG enrichment analysis and GSVA analysis were performed using the OECloud tools at https://cloud.oebiotech.com. Reactome pathway was analysed by Reactome website (https://reactome.org/). The ENCORI (https://starbase.sysu.edu.cn/) database was used to predict interactions between miRNAs and mRNAs. GSEA analysis is analyzed in the GSEA package in R. CIBERSORT analysis is performed in the Cibersort package in R. Targeted drug prediction was performed using DGIdb (https://www.dgidb.org/). Pearson correlation analysis was used for correlation analysis between FR-DEGs and immune infiltrating cells, and the AUC was used to evaluate the diagnostic efficacy of FR-DEGs in asthma. Cytoscape software was used for visualization of FR-DEGs-targeted drug networks and miRNA-miRNA networks. Statistical analysis was performed using SPSS 24.0 and GraphPad Prism 6. The measurement data were tested for normality, normally distributed data were expressed as mean ± standard deviation, and t-test for independence was used for comparison between two groups, and qualitative data were expressed as N (%); The Mann–Whitney U-test was used for comparison between two groups of non-normally distributed continuous variables. P-values less than 0.05 were statistically significant.
Results
Differentially Expressed Genes in Asthma
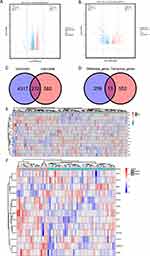
The “GEO2R” and “R/limma” packages were used to perform normalized and differential gene expression analysis on the GSE41861 and GSE43696 datasets that were taken from the GEO database. The results demonstrated that the boxplot distribution trends for the two datasets are essentially linear (Figure 2A and B). Additionally, PCA demonstrated good data reproducibility (Figure 2C and D).
According to the adjusted P value<0.05, 4588 asthma-related DEGs (2325 up-regulated and 2263 down-regulated) were screened in the GSE41861 dataset, and 854 asthma-related DEGs (400 up-regulated and 454 down-regulated) were screened in the GSE43696 dataset. Volcano plots for GSE41861 and GSE43696 datasets are shown in Figure 3A and B. Then 272 common DEGs were identified in these 2 datasets, as shown in the venn plot (Figure 3C), of which 168 were down-regulated and 104 were up-regulated (Table S2). Then, we downloaded ferroptosis using the FerrDb database and intersected them with 272 co- DEGs to obtain 13 FR-DEGs between asthma and non-asthma, as shown in the venn plot (Figure 3D). Among these 13 differential genes, 11 down-regulated genes and 2 up-regulated genes were included (Table 3). Therefore, we finally identified ADAMTS13, CBR1, ELAVL1, EPAS1, CREB5, LRRFIP1, MT1G, NOX1, NR1D2, VEGFA, ACADSB, PAQR3 and STAT3 as FR-DEGs in asthma.
 |
Table 3 13 FR-DEGs Has Been Identified as Downregulated or Upregulated in Asthma |
GO, KEGG and Reactome Pathway Enrichment Analysis of FR-DEGs
GO, KEGG and Reactome analyses were carried out to elucidate the biological functions and pathways connected to the 13 FR-DEGs. According to the findings, the GO categories was mainly enriched in surfactant homeostasis, intracellular receptor signaling pathway, monocyte differentiation, oxidoreductase activity acting on NAD(P)H, quinone or similar compound as acceptor, cellular response to vascular endothelial growth factor stimulus, angiogenesis and carbonyl reductase (NADPH) activity etc (Figure 4A and B). The KEGG pathway with the greatest enrichment were AMPK signaling pathway, pathways in cancer, HIF-1 signaling pathway, fatty acid degradation and growth hormone synthesis, etc (Figure 4C). Reactome pathway indicated that the interleukin-4 and interleukin-13 signaling, interleukin-2 family signaling and interleukin-10 signaling, regulation of gene expression by hypoxia-inducible factor, negative regulation of MAPK pathway, interleukin-7 signaling and cellular response to hypoxia were enriched (Figure 4D).
Construction of the FR-DEGs Network
We predicted the target miRNAs of FR-DEGs using the ENCORI tool. We identified 178 target miRNAs of 13 FR-DEGs. Based on the predicted 191 nodes and 370 edges, we constructed the mRNA-miRNA network by Cytoscape software (Figure 5). ADAMTS13 is regulated by 3 miRNAs, CBR1 is regulated by 11 miRNAs, ELAVL1 is regulated by 6 miRNAs, EPAS1 is regulated by 49 miRNAs, CREB5 is regulated by 31 miRNAs, LRRFIP1 is regulated by 40 miRNAs, MT1G is regulated by 7 miRNAs, NOX1 is regulated by 6 miRNAs, NR1D2 is regulated by 57 miRNAs, VEGFA is regulated by 68 miRNAs, ACADSB is regulated by 29 miRNAs, PAQR3 is regulated by 27 miRNAs and STAT3 is regulated by 36 miRNAs respectively (Table S6). Finally, using the Miranda database, mRNA-miRNA nucleic acid binding was predicted (Table S6).
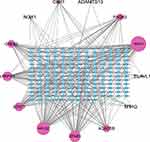 |
Figure 5 MiRNA-mRNA regulatory network of FR-DEGs. The network included 191 nodes (178 miRNAs and 13 genes) and 370 edges. |
Single‐Gene GSEA Enrichment Analysis
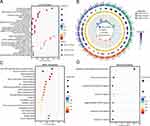
To more enrich and fully explore the role of these 13 FR-DEGs in asthma, we investigated the correlation between all genes and FR-DEGs in the GSE43696 dataset by single-gene GSEA enrichment analysis. Figure 6A–M shows the top ten enriched pathways for each FR-DEG.
By single-gene GSEA enrichment analysis, the results showed that these 13 FR-DEGs were mainly enriched in apoptosis, ferroptosis, lysosome, necroptosis, peroxisome, phagosome, proteasome, ribosome, amino acid metabolic pathway (“Glutathione metabolism”, “2-Oxocarboxylic acid metabolism”, “Alanine, aspartate and glutamate metabolism”, “Oxidative phosphorylation”, “Fatty acid metabolism”, “Porphyrin metabolism”, “Cysteine and methionine metabolism”, “Pyruvate metabolism”, “Citrate cycle (TCA cycle)”, “Arachidonic acid metabolism” and “Amino sugar and nucleotide sugar metabolism”), immune response (“Th1 and Th2 cell differentiation”, “Neutrophil extracellular trap formation”, “T cell receptor signaling pathway”, “B cell receptor signaling pathway” and“Th17 cell differentiation”) and various disease pathways (“Asthma”, “Acute myeloid leukemia”, “Influenza A”, “Gastric cancer”, “Tuberculosis” and “Hepatitis B and C”). In addition, most FR-DEGs were enriched in “JAK-STAT”, “Foxo”, “IL-17”, “NF-kappaB”, “Notch”, “PPAR”, “TGF-beta”, “Toll-like receptor”, “HIF-1”, “VEGF” and “Wnt” signaling pathways.
GSVA Enrichment Analysis
Based on the levels of each gene’s expression in combination with GSVA, we were able to identify the pathways that were differentially activated between the asthma and non-asthma groups (Figure 7A and B). The results showed that in the asthma group, the “FC-GAMMA-R-mediated-phagocytosis”, “Peroxisome”, “RNA- degradation”, “Cell-cycle”, “Long-term-potentiation”, “Regulation-of-autophagy”, “Amino-sugar-and-nucleotide-sugar-metabolism” and the signaling pathway (“VEGF”, “Calcium”, “T-cell-receptor” and “GNRH”) was activated, while the “ABC-transporters”, “Galactose-metabolism”, “Phenylalanine-metabolism”, “Glycerolipid-metabolism”, “Histidine-metabolism”, “Phenylalanine-metabolism”, and“Notch-signaling-pathway” was inhibited.
 |
Figure 7 GSVA pathway analysis. High‐ and low‐expression groups in asthma-vs-controls (A) and controls -vs- asthma (B). |
Immune Infiltration Analysis
Previous functional enrichment results showed that FR-DEGs were associated with immune response pathways. Studies had shown a strong connection between asthma and the immune microenvironment.25,26 Consequently, we next used the GSE41861 database in the Cibersort package to predict the proportion of 22 immune infiltrating cells in the immune microenvironment of asthma. The results are shown in Figure 8A, the percentages of dendritic cells activated, plasma cells, T cells CD4 memory activated and mast cells activated in asthma were higher than controls, while macrophages M2, T cells regulatory (Tregs) and T cells follicular helper were lower. Moreover, Pearson analysis suggested that plasma cells were positively and negatively correlated with ADAMTS13 (p<0.01, r=0.356) and MT1G (p<0.01, r=−0.332,) respectively, Tregs had positive and negative correlations with ELAVL1 (p<0.01, r=0.374,) and CREB5 (p<0.01, r= −0.317,) respectively, macrophages M2 had positive correlations with ADAMTS13 (p<0.01, r=0.368,) and ELAVL1 (p<0.01, r= 0.349), respectively, mast cells activated had positive and negative correlations with CBR1 (p<0.01, r=−0.379,) and NR1D2 (p<0.01, r=−0.368,) respectively, NOX1 had positive correlations with T cells CD4 memory activated (p<0.01, r=0.307)(Figure 8B; Table S7).
Targeted Drug Prediction
We predicted the targeted drugs of FR-DEGs using the DGIdb database (Table S8) and further demonstrated the FR-DEGs-drug network diagram using Cytoscape software (Figure 9). We investigated 71 drugs that target FR-DEGs, including 23 for STAT3, 5 for CBR1, 2 for ELAVL1, 3 for EPAS1, NOX1 targeted 2 drugs and VEGFA targeted 37 drugs. Unfortunately, we failed to predict the targeted drugs for ADAMTS13, CREB5, LRRFIP1, MT1G, NR1D2 and PAQR3. Moreover, the structural formulas of the 71 drugs listed above were retrieved using the DrugBank database. A total of 44 drug structures were retrieved. The structural information of the targeted drugs was revealed. The detailed results were illustrated in (Figure S1).
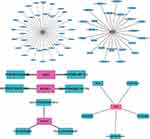 |
Figure 9 Prediction of gene‐targeted drugs (VEGFA, STAT3, NOX1, ELAVL1, EPAS1 and CBR1), The drugs may target genes through the DGIdb database. |
GSE63142 Confirmed Differentially Expression and Diagnostic Efficacy of the FR-DEGs
Finally, we verified the expression of FR-DEGs with the GSE63142 dataset. The results showed that the expression of the 10 FR-DEGs (ADAMTS13, CBR1, CREB5, ELAVL1, EPAS1, LRRFIP1, MT1G, NOX1, NR1D2 and VEGFA) were consistent with the GSE41861 and GSE43696 dataset. The expression of CBR1 (p<0.001) and CREB5 (p≤0.001) in asthma patients was greater than controls, while the ADAMTS13 (p=0.01), ELAVL1 (p=0.003), EPAS1 (p<0.001), LRRFIP1 (p=0.015), MT1G (p<0.001), NOX1 (p<0.001), NR1D2 (p<0.001) and VEGFA (p≤0.001) were lower in asthma patients (Figure 10A–J).
We created ROC curves for these 10 FR-DEGs in the GSE63142 database. The AUC of ADAMTS13 was 0.709 (95% CI: 0.595–0.823), CBR1 was 0.757 (95% CI: 0.652–0.863), CREB5 was 0.747 (95% CI: 0.634–0.861), ELAVL1 was 0.72 (95% CI: 0.601–0.839), EPAS1 was 0.722 (95% CI: 0.610–0.834), LRRFIP1 was 0.668 (95% CI: 0.546–0.790), MT1G was 0.851 (95% CI: 0.763–0.938), NOX1 was 0.812 (95% CI: 0.717–0.908), NR1D2 was 0.813 (95% CI: 0.717–0.910), and VEGFA was 0.729 (95% CI: 0.607–0.850) (Figure 11A–J). The results showed that these 10 FR-DEGs had good diagnostic roles in asthma.
Experimental Verification of Ferroptosis Genes in Asthma
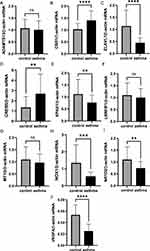
Based on the asthma patients and healthy controls, the mRNA levels of 10 FR-DEGs were then measured. The general characteristics of the asthma and control are summarized in Table 4. The results showed that the mRNA levels of CBR1 and CREB5 were elevated in asthma patients compared to healthy controls, while the mRNA levels of ELAVL1, EPAS1, NOX1, NR1D2 and VEGFA were decreased. However, the expression levels of ADAMTS13, LRRFIP1, MT1G showed no significant difference between the two groups (Figure 12A–J).
 |
Table 4 Characteristics of Asthma and Control in This Study |
Discussion
Asthma is a common and frequent disease that seriously endangers human health. In recent years, the incidence and mortality of asthma are on the rise. The main pathological changes of asthma include chronic airway inflammation, smooth muscle dysfunction, airway remodeling and mucus hypersecretion.27,28 Therefore, asthma treatment can be achieved by modulating inflammation caused by abnormalities in the immune system,15,29 restoring airway smooth muscle (ASM) function and reducing airway remodeling.30,31 Although current treatments are effective in relieving clinical symptoms in patients with asthma, a large number of patients continue to suffer from worsening or treatment-related side effects. Therefore, there is a need to investigate more effective and safe treatment options,3 provided that the molecular biological process of asthma pathogenesis must be deeply explored to lay the theoretical foundation for subsequent experimental studies. The current study found that the regulation of airway inflammation in asthma is related to ferroptosis,16,18 but studies on asthma and ferroptosis are rare. Therefore, this study explored the pathogenesis of ferroptosis in asthma using bioinformatics analysis, providing important evidence for the treatment and prevention of asthma.
Ferroptosis is a form of programmed non-apoptotic cell death caused by the accumulation of lipid peroxides and iron metabolism.4 Cell ferroptosis occurs when polyunsaturated fatty acids (PUFAs) are catalyzed by iron-containing lipoxygenases or divalent iron to form ROS and lipid peroxides, which accumulate in excess and eventually cause cell death.4 The lipid repair enzyme glutathione peroxidase 4 (GPx4) is an important protective factor of the ferroptosis process, which inhibits ferroptosis by reducing the production of lipid peroxides and ROS by attenuating lipid hydroperoxidation.32,33 Current studies have found that ferroptosis plays a new role in the pathogenesis of airway inflammation in asthma, but the specific mechanism remains to be further studied. Tang’s study found elevated airway inflammatory indicators of asthma, ROS levels, markers of ferroptosis, and lipid peroxides in mice with house dust mite induced asthma,18 suggesting that ferroptosis plays a role in the pathology of asthma. In Zhao’s study, it was demonstrated that there is a relationship between ferroptosis and autophagy in the pathogenesis of asthma, that autophagy isolates ferritin and thus increases intracellular divalent iron levels,34 and that the small scaffolding protein Phosphatidylethanolamine binding protein 1 (PEBP1) binds to iron-containing lipoxygenase and then facilitates the ferroptosis process by forming lipid peroxides. Further studies found that Ferr-1, a ferroptosis inhibitor, reduces airway inflammation in asthma by inhibiting the process of lipid peroxide production and iron release.16 Thus, it can be concluded that ferroptosis has a potential but integral role in asthma. Our study enriches the mechanism of ferroptosis in asthma through bioinformatics. The results suggest that FR-DEGs regulate asthma through oxidative stress, lysosomes, phagosomes, peroxisomes, proteasomes, autophagy, various immune response pathways, and molecular signaling pathways, but the specific mechanisms need further investigation.
The study screened out 13 FR-DEGs including ADAMTS13, CBR1, ELAVL1, EPAS1, CREB5, LRRFIP1, MT1G, NOX1, NR1D2, VEGFA, ACADSB, PAQR3 and STAT3. Among the 13 FR-DEGs s, ADAMTS13, CREB5, MT1G, and STAT3 are ferroptosis suppressor genes, while ELAVL1, EPAS1, NOX1, NR1D2, ACADSB and PAQR3 are ferroptosis driver genes. Through GO, KEGG and Reactome pathway analysis, it was finally concluded that 13 FR-DEGs were mainly enriched in oxidative stress and immune response pathways, which further confirmed that FR-DEGs may be closely associated with the pathogenesis of asthma.
The results of GSEA and GSVA analysis showed that these genes were enriched in ferroptosis, lysosomes, necrosis, autophagy, peroxisomes and phagosomes apoptosis, which were closely related to oxidative stress. Most of these genes were enriched in the immune signaling pathway (“Th1 and Th2 cell differentiation”, “Neutrophil extracellular trap formation”, “T cell receptor signaling pathway”, “B cell receptor signaling pathway” and “Th17 cell differentiation”), which is closely related to the immune response. Both oxidative stress and immune response are important pathogenesis of airway inflammation in asthma.35,36 GSVA analysis revealed that these genes promote the ferroptosis process by activating “Regulation-of-autophagy”. Xue’s study37 found that by enhancing the binding of the autophagy receptor for GPX4 protein degradation to GPX4, the autophagic degradation of GPX4 occurs, which eventually leads to ferroptosis, and the study confirmed that ferroptosis can be promoted by promoting autophagy. Zhao’s study identified a co-regulatory molecule PEBP1 of ferroptosis and autophagy, PEBP1 is also known as a rheostat between ferroptosis and autophagy, promoting ferroptosis in asthma Human airway epithelial cells (HAECs).34 The current research on ferroptosis and autophagy in asthma is very limited, and more research is needed to explore. Furthermore, by GSVA analysis, we found that these specific ferroptosis genes could promote ferroptosis through activation of ‘Peroxisome’. Peroxisomes promote ferroptosis by providing substrate PUFA-ePLs for lipid peroxidation. Peroxisome gene knockout can reduce the abundance of peroxisomes, thereby reducing the inhibitory effect of GPX4 on ferroptosis, and also promote the ferroptosis process.38 The “T cell receptor signaling pathway” is also involved in ferroptosis. Both in vivo and in vitro experiments, ferroptosis can promote the proliferation and differentiation of CD4 T cells and the production of cytokines by Th1 and Th17 cells through the T-cell receptor (TCR) signaling pathway, leading to inflammatory response.39 GPX4 can inhibit Treg ferroptosis to maintain cellular immune balance. When GPX4 is deleted, Treg induces ferroptosis under TCR/CD28 co-stimulation.40 No studies have yet investigated how ferroptosis plays a role in asthma through these pathways, and further studies are needed. The results of GSVA analysis could provide new ideas for the study of ferroptosis mechanism in asthma.
Immune homeostasis regulated by immune cells and various inflammatory factors have an important role in maintaining normal lung function, and these immune cells include T and B cells, neutrophils, mast cells, monocytes, eosinophils and dendritic cells (DC).15 Our results indicated that plasma cells, dendritic cells, mast cells and T cells CD4 memory were higher in asthma group, while macrophages M2, T cells follicular helper and Tregs were lower than the normal group by immune infiltration analysis. The result demonstrated that in asthma, the expression of CREB5 and CBR1 was high, and the expression of ELAVL1 and NR1D2 was low. Immune infiltration analysis showed that ELAVL1 was positively correlated with Tregs, CREB5 was negatively correlated with Tregs, NR1D2 was negatively correlated with mast cells, and CBR1 was positively correlated with mast cells. Consequently, ELAVL1, CREB5, NR1D2 and CBR1 may be latent targets for the treatment of asthma. Recently, it was discovered that ferroptosis is essential for immune homeostasis related to chronic inflammatory diseases.41 In neutrophil asthma, an excessive accumulation of ROS causes oxidative damage in addition to ferroptosis.20 These 13 FR-DEGs except LRRFIP1, MT1G and VEGFA were all associated with neutrophil extracellular traps (NETs), NETs formation was upregulated in asthma. As a result, ferroptosis may also modulate neutrophil resistance to pathogens by affecting NETs formation, this can lead to the development of asthma.
In our results, it was shown that the function of Tregs might be induced by activation of ELAVL1 to alleviate asthma airway inflammation. The most well-known RNA-binding protein (RBP), ELAVL1, is crucial for the biological processes of cell cycle development, immune response, inflammation, and apoptosis.42 However, according to other studies, ELAVL1 increased the levels of pro-inflammatory cytokines in CD4+T cells,43 and ELAVL1 inhibitors could improve the pro-inflammatory response,44 which seems to be contrary to our results, possibly because of the different cells selected. The cAMP response element-binding (CREB) protein family includes CREB5. CREB5 plays crucial roles in apoptosis, gene regulation and cell proliferation.45,46 Su found that Fn1 deposition in renal fibers could be regulated by targeting CREB5, which was closely related to ferroptosis.47 Additionally, in Liu’s study, CREB5 was identified as a regulator of asthma exacerbation and could be targeted to treat asthma by inhibiting CREB5.48 Therefore, we hypothesized that CREB5 may regulate asthma through the process of ferroptosis, which is consistent with our findings. NR1D2 is a nuclear hormone receptor associated with circadian clock disorder, which has attracted much attention in cancer research.49 However, there are few studies on NR1D2 in asthma, and only one study on primary HAECs used to mimic asthma with circadian symptoms found that NR1D2 exhibited rhythmic expression,50 the mechanism of which needs to be further investigated. Notably, the majority of other FR-DEGs have been documented in cancer, but no link has been found with asthma, and more researches are needed to ascertain their role in asthma.
We also used the DGIdb database to reveal drugs that might target FR-DEGs, and a total of six targeted drugs were queried (STAT3, CBR1, ELAVL1, EPAS1, NOX1, VEGFA). Among the five CBR1 target drugs retrieved, daunorubicin and doxorubicin were confirmed to be an inhibitor of CBR1. Daunorubicin is used in the treatment of leukemia, and doxorubicin is used in the treatment of acute leukemia and various cancers, but their application in asthma has not been reported. Nevertheless, a case report shows that a female patient with moderate to severe asthma who had breast cancer gradually improved her asthma symptoms after induction of chemotherapy and no longer required inhaled medication.51 Androstanolone, a steroid hormone generated by the adrenal glands, is a targeted drug for ELAVL1. Androstanolone is frequently known as the “mother hormone” since the body converts androstenedione into two steroid hormones (androgen and testosterone). It has been reported that androgen could reduce airway inflammation by inhibiting inflammatory factors produced by TH2 cells and promoting Treg cell function.52,53 Asthma airway inflammation was inhibited by androgen receptors via the MAPK1 and MAPK14 pathways.54 Testosterone could prevent the development of allergen-induced asthma by blocking mast cell activation.55 Whether our predicted gene‐targeted drugs can play a role is uncertain, therefore, the selected drugs can be prospectively studied.
Finally, we verified the expression of FR-DEGs using the GSE63142 dataset. The results showed that CBR1 and CREB5 were highly expressed, while ACADSB, ELAVL1, EPAS1, LRRFIP1, MT1G, NOX1, NR1D2 and VEGFA were low expressed in asthma patients. The results of the validation dataset were consistent with the previous conclusion. We also used the GSE63142 dataset to create ROC curves. These 10 genes were of great value in the diagnosis of asthma. Except for LRRFIP1, the AUC values of the 9 FR-DEGs were all greater than 0.7, demonstrating that 10 FR-DEGs could be used as diagnostic markers and therapeutic targets for asthma. Again, we eventually experimentally validated that 7 asthma-associated ferroptosis genes were consistent with the results predicted by bioinformatics.
Limitations
Firstly, ferroptosis was poorly studied in asthma. At present, several studies on ferroptosis in asthma has involved only a few conventional markers of ferroptosis. Although our study eventually predicted and verified 7 differentially expressed genes associated with ferroptosis in asthma, there is still insufficient experimental evidence. Therefore, we will design animal and cellular experiments for further research, which will take a long period. Secondly, this study only found that the FR-DEGs may play a role in asthma by participating in oxidative stress and immune response pathways through functional analysis. Therefore, further molecular biological experiments are needed to confirm how these ferroptosis genes play a role in oxidative stress and immune response of asthma and to further verify the mechanism of action of these ferroptosis genes in the development of asthma. Finally, the diagnostic value of these ferroptosis genes in asthma also needs to be demonstrated in larger populations, furthermore, clinical trials related to targeted ferroptosis in asthma need to be conducted to provide practical insights into the potential therapeutic role of targeted ferroptosis in asthma management.
Conclusion
Ultimately, this study screened 7 FR-DEGs in asthma by bioinformatics to provide clues for the pathogenesis of ferroptosis in asthma. These FR-DEGs have good diagnostic properties in asthma and can be used as potential biomarkers specific to asthma. These results not only identify potential biomarkers and therapeutic targets specific to asthma, but also deepen the understanding of the pathogenesis of asthma. The present study provides novel insights into the mechanistic study of ferroptosis in asthma, and the next step is to experimentally demonstrate the mechanism of action of these FR-DEGs in asthma as well as the pathways to find new therapeutic avenues for asthma.
Abbreviations
FR-DEGs, Ferroptosis-related differentially expressed genes; DEGs, Differentially expressed genes; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; GSEA, Gene Set Enrichment Analysis; GSVA, Gene Set Variation Analysis; GEO, Gene Expression Omnibus; PCA, Principal component analysis; MF, Molecular function; CC, Cellular component; BP, Biological process; ROC, Receiver operating characteristic; HAECs, Human airway epithelial cells; ASM, Airway smooth muscle; PUFAs, Polyunsaturated fatty acids; ROS, Reactive oxygen species; GPx4, Glutathione peroxidase 4; RBP, RNA-binding protein; NETs, neutrophil extracellular traps; PEBP1, Phosphatidylethanolamine binding protein 1; TCR, T-cell receptor; DC, Dendritic cells.
Data Sharing Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Ethics Committee of the second Hospital of Shanxi Medical University (approval number 2022 YX 079) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Acknowledgments
We gratefully acknowledge contributions from all authors and GEO databases.
Funding
This study was funded by key Research and Development Projects of Shanxi Province (201903D421066) and Basic Research Project of Shanxi Province (202203021211029).
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Mohan A, Ludwig A, Brehm C, Lugogo NL, Sumino K, Hanania NA. Revisiting mild asthma: current knowledge and future needs. Chest. 2022;161(1):26–39. doi:10.1016/j.chest.2021.09.004
2. Global Initiative for Asthma. Global Strategy for Asthma Management and Prevention, 2022 (GINA, 2022). Available from: https://ginasthma.org/.
3. Rupani H, Fong WCG, Kyyaly A, Kurukulaaratchy RJ. Recent insights into the management of inflammation in asthma. J Inflamm Res. 2021;14:4371–4397. doi:10.2147/JIR.S295038
4. Dixon SJ, Lemberg KM, Lamprecht MR, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. doi:10.1016/j.cell.2012.03.042
5. Angeli JPF, Shah R, Pratt DA, Conrad M. Ferroptosis inhibition: mechanisms and opportunities. Trends Pharmacol Sci. 2017;38(5):489–498. doi:10.1016/j.tips.2017.02.005
6. Li J, Cao F, Yin HL, et al. Ferroptosis: past, present and future. Cell Death Dis. 2020;11(2):88. doi:10.1038/s41419-020-2298-2
7. Brown CW, Amante JJ, Chhoy P, et al. Prominin2 drives ferroptosis resistance by stimulating iron export. Dev Cell. 2019;51(5):575–586.e4. doi:10.1016/j.devcel.2019.10.007
8. Yang WS, Kim KJ, Gaschler MM, Patel M, Shchepinov MS, Stockwell BR. Peroxidation of polyunsaturated fatty acids by lipoxygenases drives ferroptosis. Proc Natl Acad Sci U S A. 2016;113(34):E4966–E4975. doi:10.1073/pnas.1603244113
9. Dixon SJ, Patel DN, Welsch M, et al. Pharmacological inhibition of cystine-glutamate exchange induces endoplasmic reticulum stress and ferroptosis. Elife. 2014;3:e02523. doi:10.7554/eLife.02523
10. Ajoolabady A, Aslkhodapasandhokmabad H, Libby P, et al. Ferritinophagy and ferroptosis in the management of metabolic diseases. Trends Endocrinol Metab. 2021;32(7):444–462. doi:10.1016/j.tem.2021.04.010
11. Fang X, Wang H, Han D, et al. Ferroptosis as a target for protection against cardiomyopathy. Proc Natl Acad Sci U S A. 2019;116(7):2672–2680. doi:10.1073/pnas.1821022116
12. Mahoney-Sanchez L, Bouchaoui H, Boussaad I, et al. Alpha synuclein determines ferroptosis sensitivity in dopaminergic neurons via modulation of ether-phospholipid membrane composition. Cell Rep. 2022;40(8):111231. doi:10.1016/j.celrep.2022.111231
13. Xie J, Zhang T, Li P, Wang D, Liu T, Xu S. Dihydromyricetin attenuates cerebral ischemia reperfusion injury by inhibiting SPHK1/mTOR signaling and targeting ferroptosis. Drug Des Devel Ther. 2022;16:3071–3085. doi:10.2147/DDDT.S378786
14. Walters R, Mousa SA. Modulations of ferroptosis in lung cancer therapy. Expert Opin Ther Targets. 2022;26(2):133–143. doi:10.1080/14728222.2022.2032651
15. Lv X, Dong M, Tang W, et al. Ferroptosis, novel therapeutics in asthma. Biomed Pharmacother. 2022;153:113516. doi:10.1016/j.biopha.2022.113516
16. Yang N, Shang Y. Ferrostatin-1 and 3-methyladenine ameliorate ferroptosis in OVA-induced asthma model and in IL-13-challenged BEAS-2B cells. Oxid Med Cell Longev. 2022;2022:9657933. doi:10.1155/2022/9657933
17. Han F, Li S, Yang Y, Bai Z. Interleukin-6 promotes ferroptosis in bronchial epithelial cells by inducing reactive oxygen species-dependent lipid peroxidation and disrupting iron homeostasis. Bioengineered. 2021;12(1):5279–5288. doi:10.1080/21655979.2021.1964158
18. Tang W, Dong M, Teng F, et al. Environmental allergens house dust mite-induced asthma is associated with ferroptosis in the lungs. Exp Ther Med. 2021;22(6):1483. doi:10.3892/etm.2021.10918
19. Zeng Z, Huang H, Zhang J, et al. HDM induce airway epithelial cell ferroptosis and promote inflammation by activating ferritinophagy in asthma. FASEB J. 2022;36(6):e22359. doi:10.1096/fj.202101977RR
20. Bao C, Liu C, Liu Q, et al. Liproxstatin-1 alleviates LPS/IL-13-induced bronchial epithelial cell injury and neutrophilic asthma in mice by inhibiting ferroptosis. Int Immunopharmacol. 2022;109:108770. doi:10.1016/j.intimp.2022.108770
21. Wang Y, Wan R, Peng W, Zhao X, Bai W, Hu C. Quercetin alleviates ferroptosis accompanied by reducing M1 macrophage polarization during neutrophilic airway inflammation. Eur J Pharmacol. 2023;938:175407. doi:10.1016/j.ejphar.2022.175407
22. Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14(1):7. doi:10.1186/1471-2105-14-7
23. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi:10.1038/nmeth.3337
24. Freshour SL, Kiwala S, Cotto KC, et al. Integration of the drug-gene interaction database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res. 2021;49(D1):D1144–D1151. doi:10.1093/nar/gkaa1084
25. Lin Q, Ni H, Zheng Z, Zhong J, Nie H. Cross-talk of four types of RNA modification writers defines the immune microenvironment in severe asthma. Ann N Y Acad Sci. 2022;1514(1):93–103. doi:10.1111/nyas.14782
26. Min Z, Zeng Y, Zhu T, et al. Lipopolysaccharide-activated bone marrow-derived dendritic cells suppress allergic airway inflammation by ameliorating the immune microenvironment. Front Immunol. 2021;12:595369. doi:10.3389/fimmu.2021.595369
27. Ntontsi P, Photiades A, Zervas E, Xanthou G, Samitas K. Genetics and Epigenetics in Asthma. Int J Mol Sci. 2021;22(5):2412. doi:10.3390/ijms22052412
28. Bønnelykke K, Ober C. Leveraging gene-environment interactions and endotypes for asthma gene discovery. J Allergy Clin Immunol. 2016;137(3):667–679. doi:10.1016/j.jaci.2016.01.006
29. Wen X, Liu HX, Chen LZ, et al. Asthma susceptibility in prenatal nicotine-exposed mice attributed to β-catenin increase during CD4+ T cell development. Ecotoxicol Environ Saf. 2022;238:113572. doi:10.1016/j.ecoenv.2022.113572
30. Nguyen LP, Al-Sawalha NA, Parra S, et al. β2-Adrenoceptor signaling in airway epithelial cells promotes eosinophilic inflammation, mucous metaplasia, and airway contractility. Proc Natl Acad Sci U S A. 2017;114(43):E9163–E9171. doi:10.1073/pnas.1710196114
31. Fayon M, Rebola M, Berger P, et al. Increased secretion of leukemia inhibitory factor by immature airway smooth muscle cells enhances intracellular signaling and airway contractility. Am J Physiol Lung Cell Mol Physiol. 2006;291(2):L244–L251. doi:10.1152/ajplung.00474.2005
32. Friedmann Angeli JP, Schneider M, Proneth B, et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol. 2014;16(12):1180–1191. doi:10.1038/ncb3064
33. Imai H, Hirao F, Sakamoto T, et al. Early embryonic lethality caused by targeted disruption of the mouse PHGPx gene. Biochem Biophys Res Commun. 2003;305(2):278–286. doi:10.1016/s0006-291x(03)00734-4
34. Zhao J, Dar HH, Deng Y, et al. PEBP1 acts as a rheostat between prosurvival autophagy and ferroptotic death in asthmatic epithelial cells. Proc Natl Acad Sci U S A. 2020;117(25):14376–14385. doi:10.1073/pnas.1921618117
35. Michaeloudes C, Abubakar-Waziri H, Lakhdar R, et al. Molecular mechanisms of oxidative stress in asthma. Mol Aspects Med. 2022:85. doi:10.1016/j.mam.2021.101026
36. Holgate ST. Innate and adaptive immune responses in asthma. Nat Med. 2012;18(5):673–683. doi:10.1038/nm.2731
37. Xue Q, Yan D, Chen X, et al. Copper-dependent autophagic degradation of GPX4 drives ferroptosis. Autophagy. 2023;19(7):1982–1996. doi:10.1080/15548627.2023.2165323
38. Zou Y, Henry WS, Ricq EL, et al. Plasticity of ether lipids promotes ferroptosis susceptibility and evasion. Nature. 2020;585(7826):603–608. doi:10.1038/s41586-020-2732-8
39. Luoqian J, Yang W, Ding X, et al. Ferroptosis promotes T-cell activation-induced neurodegeneration in multiple sclerosis. Cell Mol Immunol. 2022;19(8):913–924. doi:10.1038/s41423-022-00883-0
40. Xu C, Sun S, Johnson T, et al. The glutathione peroxidase Gpx4 prevents lipid peroxidation and ferroptosis to sustain Treg cell activation and suppression of antitumor immunity. Cell Rep. 2021;35(11):109235. doi:10.1016/j.celrep.2021.109235
41. Wu Y, Chen H, Xuan N, et al. Induction of ferroptosis-like cell death of eosinophils exerts synergistic effects with glucocorticoids in allergic airway inflammation. Thorax. 2020;75(11):918–927. doi:10.1136/thoraxjnl-2020-214764
42. Lin Z, Song J, Gao Y, et al. Hypoxia-induced HIF-1α/lncRNA-PMAN inhibits ferroptosis by promoting the cytoplasmic translocation of ELAVL1 in peritoneal dissemination from gastric cancer. Redox Biol. 2022;52:102312. doi:10.1016/j.redox.2022.102312
43. Techasintana P, Ellis JS, Glascock J, et al. The RNA-binding protein HuR posttranscriptionally regulates IL-2 homeostasis and CD4+ Th2 differentiation. Immunohorizons. 2017;1(6):109–123. doi:10.4049/immunohorizons.1700017
44. Fattahi F, Ellis JS, Sylvester M, et al. HuR-targeted inhibition impairs Th2 proinflammatory responses in asthmatic CD4+ T cells. J Immunol. 2022;208(1):38–48. doi:10.4049/jimmunol.2100635
45. Rezaee F, Harford TJ, Linfield DT, et al. cAMP-dependent activation of protein kinase A attenuates respiratory syncytial virus-induced human airway epithelial barrier disruption. PLoS One. 2017;12(7):e0181876. doi:10.1371/journal.pone.0181876
46. Koschinski A, Zaccolo M. Activation of PKA in cell requires higher concentration of cAMP than in vitro: implications for compartmentalization of cAMP signalling. Sci Rep. 2017;7(1):14090. doi:10.1038/s41598-017-13021-y
47. Su H, Xie J, Wen L, et al. LncRNA Gas5 regulates Fn1 deposition via Creb5 in renal fibrosis. Epigenomics. 2021;13(9):699–713. doi:10.2217/epi-2020-0449
48. L X, Kg N, La S, et al. Single-cell RNA transcriptomic analysis identifies Creb5 and CD11b-DCs as regulator of asthma exacerbations. Mucosal Immunol. 2022;15(6). doi:10.1038/s41385-022-00556-1
49. Ercolani L, Ferrari A, De Mei C, Parodi C, Wade M, Grimaldi B. Circadian clock: time for novel anticancer strategies? Pharmacol Res. 2015;100:288–295. doi:10.1016/j.phrs.2015.08.008
50. Powell WT, Rich LM, Vanderwall ER, White MP, Debley JS. Temperature synchronisation of circadian rhythms in primary human airway epithelial cells from children. BMJ Open Respir Res. 2022;9(1):e001319. doi:10.1136/bmjresp-2022-001319
51. Palmieri C, Gillmore R, Menzies-Gow A, et al. Resolution of late-onset asthma following high-dose chemotherapy. Bone Marrow Transplant. 2003;32(8):847–848. doi:10.1038/sj.bmt.1704268
52. Xia T, Ma J, Sun Y, Sun Y. Androgen receptor suppresses inflammatory response of airway epithelial cells in allergic asthma through MAPK1 and MAPK14. Hum Exp Toxicol. 2022;41:9603271221121320. doi:10.1177/09603271221121320
53. Ejima A, Abe S, Shimba A, et al. Androgens alleviate allergic airway inflammation by suppressing cytokine production in Th2 cells. J Immunol. 2022;209(6):1083–1094. doi:10.4049/jimmunol.2200294
54. Gandhi VD, Cephus JY, Norlander AE, et al. Androgen receptor signaling promotes Treg suppressive function during allergic airway inflammation. J Clin Invest. 2022;132(4):e153397. doi:10.1172/JCI153397
55. Cerqua I, Terlizzi M, Bilancia R, et al. 5α-dihydrotestosterone abrogates sex bias in asthma like features in the mouse. Pharmacol Res. 2020;158:104905. doi:10.1016/j.phrs.2020.104905
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.