Back to Journals » International Journal of Nanomedicine » Volume 18
Exosomes Derived from E2F1–/– Adipose-Derived Stem Cells Promote Skin Wound Healing via miR-130b-5p/TGFBR3 Axis
Authors Yu H, Wu Y, Zhang B, Xiong M, Yi Y, Zhang Q , Wu M
Received 21 July 2023
Accepted for publication 21 October 2023
Published 3 November 2023 Volume 2023:18 Pages 6275—6292
DOI https://doi.org/10.2147/IJN.S431725
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor R.D.K. Misra
Honghao Yu,* Yiping Wu,* Boyu Zhang, Mingchen Xiong, Yi Yi, Qi Zhang, Min Wu
Department of Plastic and Cosmetic Surgery, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, 430030, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Qi Zhang; Min Wu, Tel +86 15951072198 ; +86 13554254796, Email [email protected]; [email protected]
Background: Skin wound is a widespread health problem and brings extraordinary burdens to patients. Exosomes derived from adipose-derived stem cells (ADSC-Exos) are considered promising strategies for repairing skin wounds. E2F1 is a member of the E2F family of transcription factors involved in cell growth and apoptosis. E2F1 deficiency in mice enhances wound healing by improving collagen deposition and angiogenesis. Additionally, E2F1 can regulate the transcription and paracrine activity of multiple miRNAs, which will inevitably reshape the paracrine expression profile of ADSC-Exos. This study aimed to investigate the impact of transcription factor E2F1 deficiency on the functions of ADSC-Exos in promoting wound healing.
Methods: First, we obtained ADSCs from subcutaneous adipose tissues of WT and E2F1–/– C57BL/6 mice and separated their exosomes, denoted as ADSCWT-Exos and ADSCE2F1-/--Exos. The wound healing effects of ADSCWT-Exos and ADSCE2F1-/--Exos in full-thickness skin wound models were investigated by wound images, H&E staining, and immunohistochemical staining. For the in vitro study, the abilities of ADSCWT-Exos and ADSCE2F1-/--Exos to promote cell activities, collagen formation, and angiogenesis were evaluated. The potential mechanism by which ADSCE2F1-/--Exos promote wound healing was determined by miRNA sequencing, ChIP‒qPCR, and dual-luciferase assays.
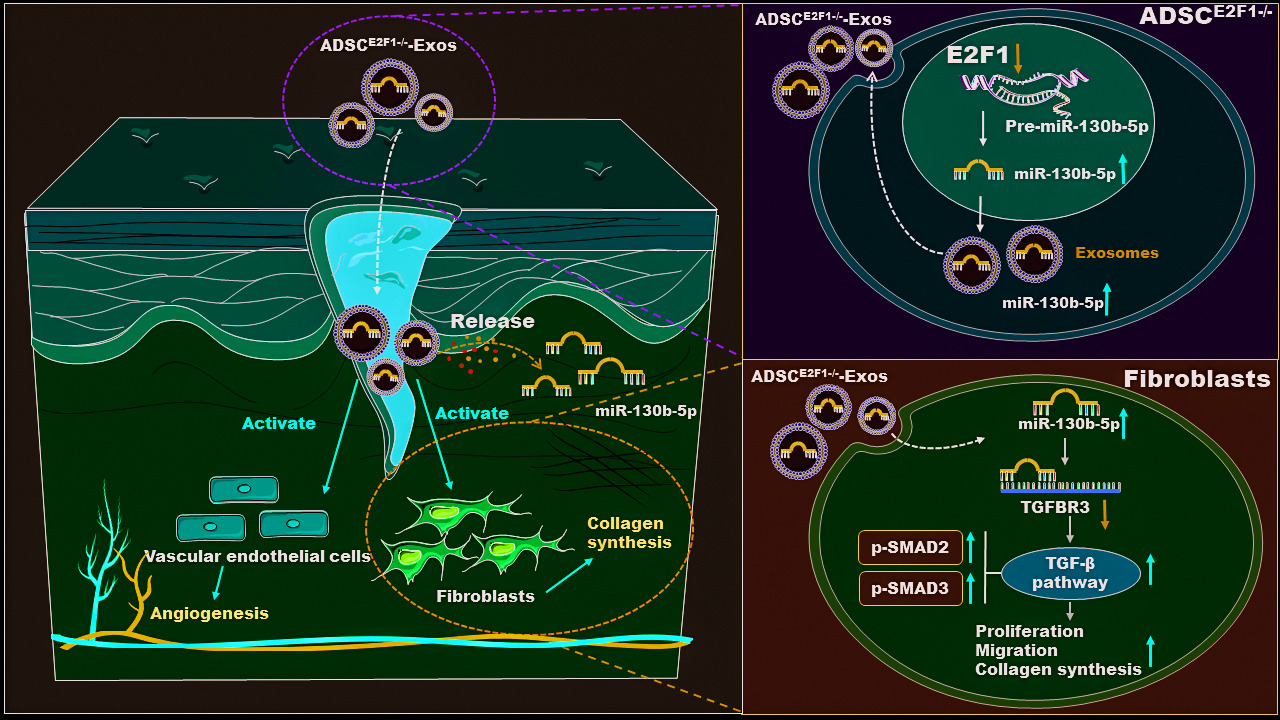
Results: ADSCE2F1-/--Exos accelerated wound healing by promoting collagen formation and angiogenesis. As a result, compared with the lower wound healing rate of 30.5% within 7 days in the control group and 42.3% in the ADSCWT-Exo group, ADSCE2F1-/--Exos significantly increased the wound healing rate to 72.5%. In vitro, ADSCE2F1-/--Exos activated the function of fibroblasts and vascular endothelial cells. The loss of E2F1 promoted miR-130b-5p expression in ADSCE2F1-/--Exos through transcriptional regulation. MiRNA high-throughput sequencing identified 12 differently expressed miRNAs between ADSCE2F1-/- and ADSCWT. ADSCE2F1-/--Exos enhanced fibroblast activities via the miR-130b-5p/TGFBR3 axis and TGF-β activation.
Conclusion: Our results indicated that ADSCE2F1-/--Exos effectively promoted wound healing by regulating the miR-130b-5p/TGFBR3 axis, thus providing a novel strategy of gene-engineered stem cell exosomes for accelerating wound healing.
Keywords: wound healing, adipose-derived stem cells, exosome, E2F1, miR-130b-5p
Graphical Abstract:
Introduction
With the increasing occurrence of senile and metabolic diseases, the number of patients with chronic and refractory wounds is increasing year by year.1 Chronic and refractory wounds seriously affect the living quality of patients and and place a heavy economic burden on families and society.2 Wound healing is an orderly and highly coordinated process, which is consisted of four overlapping phases, including hemostasis, inflammation, proliferation, and tissue remodeling.3 During these phases, multiple different cell types dynamically interact with each other and play specific roles in mediating the wound healing process.4 During the hemostasis phase, injured blood vessels constrict, platelets activate to form fibrin clots, and immune cells are recruited to the wound site. In the inflammatory phase, neutrophils and macrophages clear bacteria, and secrete cytokines and growth factors to initiate the beginning of proliferation phase. In the proliferation stage, vascular endothelial cells participate in the formation of new blood vessels, fibroblasts produce collagen and remodel the extracellular matrix (ECM), and keratinocytes migrate to re-epithelialize. In the remodeling phase, cells in the granulation tissue undergo apoptosis, and macrophages break down excessive ECM and apoptotic cells.5 Therefore, regulating and intervening with cells and factors in these 4 processes is an effective strategy to promote wound healing.
Adipose-derived stem cells (ADSCs) are considered to be one of the most ideal stem cells to promote wound healing and tissue regeneration due to their rich and convenient sources and multi-differentiation potential.6 However, the strict storage conditions, allogeneic use safety, and ethics problem limit the wide clinical application of ADSCs.7 The role of ADSCs in promoting tissue damage repair and regeneration is primarily dependent on stemness differentiation capacity and strong paracrine effects.8 Studies have shown that ADSCs can significantly enhance cell proliferation, angiogenesis, collagen synthesis, and tissue regeneration via releasing multiple paracrine exosomes, cytokines, and growth factors.9 Exosomes are defined as extracellular vesicles with a lipid bilayer structure of 30 to 150 nm in diameter, which encapsulate proteins, nucleic acids (DNA, mRNA, non-coding RNA), lipids, and small molecular metabolites.10 Exosomes in general can be secreted by almost all cells, and their biomarkers and cargoes are a mapping of their parental cells, implying that ADSC-Exos inherits the functions of parental ADSCs to some extent.11 ADSC-Exos have been verified to improve osteoarthritis, promote the effectiveness of fat transplantation, and accelerate skin wound regeneration.12–14 It is worth noting that the paracrine function and pro-wound healing capacity of ADSCs can be more effectively improved by pharmacological intervention, hypoxic culture, and genetic engineering editing.15 For example, hypoxic ADSC-derived exosomes promoted diabetic wound healing more effectively in comparison to the control exosomes.16 Besides, exosomes derived from SIRT1-overexpressing ADSCs improved the protective effects in acute myocardial infarction via inducing vasculogenesis and decreasing inflammation.17 Therefore, exosomes from genetically modified ADSCs are a promising strategy for wound regeneration.
E2F1 is a member of the E2F family of transcription factors and is mainly involved in the regulation of cell growth and apoptosis.18 In our previous study, we investigated the role of E2F1 in different diseases and found that E2F1 deficiency in mice not only improved cardiac performance after myocardial infarction but also enhanced wound healing in a mouse wound model.19,20 Besides, we also emphasized E2F1 deficiency could improve the ability of ADSC paracrine to promote wound healing.21 Exosomal microRNAs (miRNAs) are considered to be important paracrine substances for ADSCs in promoting wound healing.22 E2F1 can regulate the transcription of multiple miRNAs. For instance, Wang et al suggested that E2F1 activates miR-421 expression to induce mitochondrial fragmentation and apoptosis in cardiomyocytes.23 This suggests that E2F1 may interfere with the secretory expression and function of ADSCs by regulating specific miRNA populations. These regulatory effects of E2F1 will inevitably reshape the paracrine expression profile of ADSC-Exos.
Therefore, in this study, we aimed to explore the regulatory role of E2F1 on the secretory function of ADSC-Exos and the potential role and mechanism of ADSC-Exos in promoting wound healing. Firstly, we established mouse skin wound models to explore the wound healing effect of ADSCE2F1-/--derived exosome (ADSCE2F1-/--Exos). Then, we evaluated the impact of ADSCE2F1-/--Exos on the function of fibroblasts and vascular endothelial cells in vitro, including proliferation, migration, collagen synthesis, and angiogenesis. Besides, we performed miRNA high-throughput sequencing to identify the differently expressed miRNAs in ADSCE2F1-/--Exos and explored the function of the miRNAs in wound healing. The successful implementation of this method will contribute to developing novel ADSC-exo-based strategies for promoting wound healing.
Materials and Methods
Generation of E2F1–/– Mice and Acquirement of ADSCs
E2F1–/– C57BL/6 mice were obtained from the Jackson Laboratory (Bar Harbor, ME, USA), and were bred, maintained, and operated in the Animal Experimental Center of Tongji Hospital, Huazhong University of Science and Technology. ADSCs were isolated from subcutaneous adipose tissues in the inguinal region of E2F1–/– and WT C57BL/6 mice by a previously published protocol.24 All animal experiments were approved by the Animal Care Committee of Tongji Medical College (TJH-201909006). For identification of ADSCs, the phenotypic markers, including CD105, CD90, CD29, CD44, CD31, and CD34 of ADSCs were analyzed by flow cytometry, and tri-lineage differentiation capacity was verified by Alizarin red staining, Oil-red O staining, and Alcian blue staining.
Cell Culture and Transfection
ADSCs were cultured in DMEM (Gibco, USA) containing 10% fetal bovine serum (FBS) (Gibco, USA) and 1% penicillin-streptomycin solution (Gibco, USA), and were placed in an incubator with 5% CO2 at 37 °C. ADSCs from passage 3 to 6 were used for the following experiments. The mouse embryonic fibroblast NIH-3T3 cell line, and mouse vascular endothelial cell C166 cell line were obtained from the Cell Bank of the Chinese Academy of Sciences. NIH-3T3 cells and C166 cells were cultured with DMEM (Gibco, USA) containing 10% FBS (Gibco, USA) and 1% penicillin-streptomycin solution (Gibco, USA).
NIH-3T3 cells for transfection were seeded in 6-well plates and incubated to 40–50% confluence. The small interfering RNAs (siRNA) for TGFBR3, miR-130b-5p mimics, miR-130b-5p inhibitors, TGFBR3 overexpression plasmids, and the corresponding negative controls were designed and synthesized by Tsingke Biotech (Beijing, China). Transfection was conducted with Lipofectamine 3000 Transfection Reagent (Invitrogen, USA) according to the manufacturer’s instructions.
Isolation and Characterization of ADSC-Exos
ADSCs from passages 3 to 6 were cultured with DMEM (Gibco, USA) containing 10% exosome-free serum (Gibco, USA) and 1% penicillin-streptomycin solution (Gibco, USA). The supernatant was centrifuged at 2000 g for 10 min and at 10,000 g for 30 min to remove the dead cells, debris, and other large vesicles. Then the supernatant was centrifuged at 145,000 g for 70 min with Optima XPN-100 ultracentrifuge (Beckman, USA). The supernatant was removed and the exosome pellet was washed with PBS 3 times. Exosomes were filtered through a 0.22 μm filter membrane and stored at −80 °C. The protein quantitation of exosomes was determined by the BCA protein assay kit (Biosharp, China). The expression of exosome markers CD63, CD9, and HSP70 were determined by Western blot, and the dimensions and ultrastructure of purified exosomes were analysed with NanoSight (Malvern Instruments, China) and transmission electron microscope (TEM, Hitachi, Japan).
Mouse Skin Wound Model and Treatment
A total of 45 C57BL/6 mice (8 w, male) were anaesthetized with pentobarbital sodium (60 mg/kg). After hair removal and sterilization, a full-thickness skin wound was performed on the middle of mouse back with an 8 mm biopsy needle (Kai Medical, Germany). Then, 45 mice were randomly divided into 3 groups: Control (100 μL PBS) group, ADSCWT-derived exosomes (ADSCWT-Exos) (1 μg/μL in 100 μL PBS) group, and ADSCE2F1-/--Exos (1 μg/μL in 100 μL PBS) group. For treatment, 80 μL PBS suspensions were subcutaneously injected into 4 sites of the wound edge, and 20 μL was directly applied onto the middle sites of the wound. The wound areas were photographed and measured on day 0, 3, 7, and 14 post-treatment.
Histological Evaluation
The skin tissue samples, including the wound area and 2 mm surrounding wound edge, were collected for histological evaluation. The skin tissues were fixed and embedded in paraffin. Tissue sections with 4 μm thick were stained with hematoxylin and eosin (H&E) to assess the percentage of epithelization. Masson trichrome staining was performed to evaluate the degree of collagen maturity. For immunofluorescence (IF) staining, the sections were incubated with anti-CD31 antibodies (1:100, Boster Bio, China) and anti-α-SMA antibodies (1:50, Abcam, UK) at 4 °C overnight. After washing 3 times with TBST, the sections were incubated with secondary antibodies (Proteintech, China) and DAPI (Sigma-Aldrich, USA). Images were captured with a fluorescence microscope (Olympus, Japan). For immunohistochemistry (IHC) staining, the sections were incubated with anti-TGF-β1 antibodies (1:200, Bioss, China) at 4 °C overnight. Then the sections were washed with TBST solution and incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies (Proteintech, China) for 1 h. The sections were finally visualized with DAB Peroxidase Substrate Kit (Maxin, China). The images were captured with the SOPTOP CX40 microscope (China).
Cell Proliferation Assay
The 5-Ethynyl-20-Deoxyuridine (EdU) cell proliferation kit (RiboBio, China) was used to measure the proliferation ability of C166 and NIH-3T3 cells. After treatment, the cells were incubated with 50 μM EdU for 3 h. Then the cells were fixed and incubated with an Apollo reaction cocktail and DAPI staining solution. The ratio of EdU-positive cells to the total number of cells was used to measure the proliferation ability of cells.25 All the samples were prepared in triplicate.
Cell Migration Assay
Wound healing and transwell migration assay were used to assess the migration of C166 and NIH-3T3 cells. For wound healing assay, after the cells were incubated to 80–90% confluence, the cell layers were gently scratched with the 200 μL pipette tip. Afterwards, the cell layers were washed with PBS 3 times and incubated in serum-free DMEM/F12 medium for 24 h. Wound images at 0 h and 24 h were captured by SOPTOP CX40 microscope (China). The horizontal distance of migrated cells from wound edge was calculated by ImageJ software (NIH, USA).
For transwell migration assay, 5×104 cells/well were resuspended in 200 μL serum-free DMEM/F12 medium and were seeded into the upper cell chamber of transwell migration chambers (8 μm size, Corning, USA). Meanwhile, 500 μL of DMEM/F12 medium containing 20% FBS was placed in the bottom cell chamber as an attractant. After 24 h, the migrated cells of the lower surface of upper chamber were fixed and stained with 0.5% crystal violet. The number of migrated cells was calculated by ImageJ software (NIH, USA).
Tube Formation Assay
Matrigel (BD Biosciences, USA) was used for the tube formation assay. Thawed matrigel was spread on a 48-well plate for solidification. C166 cells were seeded onto matrigel and treated with ADSCWT-Exos (50 μg/mL) or ADSCE2F1-/--Exos (50 μg/ mL) for 24 h at 37 °C. Tube formation images were observed with a SOPTOP CX40 microscope.
MiRNA High-Throughput Sequence
The TRIzol reagent kit (Invitrogen, USA) was used to obtain the total RNA of ADSCWT and ADSCE2F1-/-. Then the total RNA was submitted in triplicate to Berry Genomics (Beijing, China) for construction and sequencing of the RNA-seq library on HiSeq/MiSeq (Illumina). R language R x64 4.0.5 was used to analyse data and create clustered heat map and volcano plots.
Total RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
The TRIzol reagent kit (Invitrogen, USA) was used to isolate the total RNA. NanoDrop 2000 spectrophotometer (Thermo, USA) was used to determine the integrity and concentration of RNA. Then the RNA was reversed into cDNA with miRNA 1st Strand cDNA Synthesis Kit or 1st Strand cDNA Synthesis Kit (Vazyme, China). The qRT-PCR analysis was carried out with SYBR Green Master Mix (Vazyme, China). All primer sequences used for qRT-PCR were summarized in Table S1. β-actin was used as an internal reference for mRNA, and U6 was used as an internal reference for miRNA.
Western Blot Analysis
The total proteins were extracted by IP lysis buffer (Servicebio, China) containing phosphatase inhibitors and PMSF, and then their concentrations were determined by a BCA protein assay kit (Biosharp, China). The obtained proteins were separated by SDS-PAGE and then were electrotransferred to PVDF membranes (Bio-Rad, USA). Next, the membranes were blocked with 5% bovine serum albumin (BSA) for 1 h and incubated with primary antibody overnight at 4 °C. Subsequently, the membranes were washed with TBST for 3 times and incubated with HRP-conjugated secondary antibodies for 1 h at room temperature. Finally, the ECL assay kit (Yeasen, USA) was applied to visualize the protein bands. The main antibodies used for Western blot analysis were summarized in Table S2.
Chromatin Immunoprecipitation (ChIP) Assay
ADSCs were washed with PBS and incubated for 10 min with 1% formaldehyde at room temperature. Next, ADSCs were lysed with ChIP Lysis Buffer and sonicated into chromatin fragments at 500–800 bp. Then, the samples were incubated with 5 μL of anti-E2F1 antibody and vibrated overnight at 4 °C. Protein G MagBeads (Thermo Fisher, USA) was used to capture the immunoprecipitates for 4 h. Then, immunoprecipitates were eluted from the Protein G MagBeads with ChIP Elution Buffer and digested by protease K. The DNA fragments in the buffer were purified with a DNA Purification Kit (Cwbiotech, China). Purified DNA was used for qPCR and qRT-PCR. For analyzing E2F1-binding to the miR-130b-5p promoter region, the primer sequences were as follows: forward: 5’-ATCCATGGTTGAGCTTCCCG-3’; and reverse: 5’-TAGTGCAACCTCGTCAGAGC-3’.
Fluorescence in situ Hybridization (FISH)
Cy5-labelled miR-130b-5p probes were designed and synthesized by Powerful Biology (Wuhan, China). In brief, NIH-3T3 cells were washed with PBS, fixed with 4% formaldehyde solution for 15 min at room temperature, and incubated with 0.5% Triton X-100 solution at 4 °C for 5 min. Cy5-labelled miR-130b-5p was used for situ hybridization overnight in the dark. Images were captured with a fluorescence microscope. The probe sequence of miR-130b-5p were as follows: 5’-AGTAGTGCAACAGGGAAAGAGT-3’.
Dual-Luciferase Activity Assay
The recombinant luciferase reporter plasmids of pmirGLO-TGFBR3-wt and pmirGLO- TGFBR3-mut were designed and synthesized by Tsingke (Beijing, China). After HEK293T cells reached a confluency of 40–50%, TGFBR3 plasmid together with the miR-130b-5p mimic or NC-mimic were transfected into cells via Lipofectamine 3000 Transfection Reagent (Invitrogen, USA). Then, firefly luciferase activity and Rluc activity were determined with a dual-luciferase Reporter Gene Assay Kit (Yeasen, China). Relative luciferase activity was the ratio of firefly luciferase activity to Rluc activity.26
Statistical Analysis
All the statistical analyses of the experimental data were conducted with GraphPad Prism 9.0 (GraphPad, USA). Data were obtained from at least three replicate assays, reported as mean ± standard deviation (SD). The Student’s t-test was adopted to determine the differences between the two groups. The One-way ANOVA was used to determine the differences among three or more groups. P-value < 0.05 was regarded as statistically significant.
Results
Characterization of ADSCs and Exosomes
The identification of ADSCs was carried out with optical images, tri-lineage differentiation capacity, and flow cytometry. Images captured by microscope showed that ADSCs adhered to the culture plate and presented a spindle-like morphology (Figure 1A). Alizarin red staining, Oil-red O staining, and Alcian blue staining images identified the osteogenesis, adipogenesis, and chondrogenesis capacity of ADSCs after differentiation induction (Figure 1B). Flow cytometry results showed that ADSCs were positive for the mesenchymal stromal cell (MSC) surface markers CD105, CD90, CD29, and CD44, and negative for hematopoietic lineage marker CD34 and the endothelial marker CD31 (Figure 1C). TEM, dynamic light scattering (DLS) analysis, and Western blot analysis were performed to identify the exosomes derived from ADSCs. TEM revealed that ADSCE2F1-/--Exos and ADSCWT-Exos were cup- or sphere-shaped vesicles, and DLS analysis showed that exosomes had a diameter of approximately 100 nm (Figure 1D). Western blot analysis showed that ADSCE2F1-/--Exos and ADSCWT-Exos were positive for the exosome-specific biomarkers CD63, CD9, and HSP70 (Figure 1E).
ADSCE2F1-/--Exos Improved Wound Healing in vivo
To evaluate the effect of ADSC E2F1-/--Exos on wound healing, we established the full-thickness skin wounds on C57BL/6 mice. The process of in vivo experience was presented in Figure 2A. The wound healing rate of ADSCE2F1-/--Exos group was significantly increased compared to that of the Control group and ADSCWT-Exos group at day 7, and 14 post-treatment (Figure 2B-D). Additionally, H&E staining showed that the degree of reepithelialization and cuticle coverage was improved in skin wounds of ADSCE2F1-/--Exo group. In addition, the regeneration of skin appendages in ADSCE2F1-/--Exo group, such as hair follicles, was better than the Control group and ADSCWT-Exo group (Figure 2E). Masson trichrome staining showed that orderly collagen deposits were formed in ADSCWT-Exo group and ADSCE2F1-/--Exo group at 14 days, but collagen fibers in the ADSCE2F1-/--Exo group were significantly thicker than ADSCWT-Exo group (Figure 2F). The IF images of CD31 and α-SMA confirmed that the amount of new blood vessels was significantly increased in ADSCWT-Exo group and ADSCE2F1-/--Exo group compared with the Control group. And the new blood vessels of ADSCE2F1-/--Exo group was greater than ADSCWT-Exo group (Figure 2G). For DAPI staining, the results revealed no significant difference in the cell count within the wound tissue among the three groups. These results suggested that ADSCE2F1-/--Exos could improve wound healing by promoting collagen formation and angiogenesis.
ADSCE2F1-/--Exos Promoted Collagen Formation and Angiogenesis in vitro
The wound healing assay demonstrated that NIH-3T3 cells treated with ADSCE2F1-/--Exo migrated more to the scratch area in comparison with the Control group and ADSCWT-Exo group (Figure 3A). Similarly, in transwell assay, the number of migrated cells in ADSCE2F1-/--Exo group was larger than that in the other two groups (Figure 3B). EdU assays proved that ADSCE2F1-/--Exo significantly promoted the proliferation of fibroblasts (Figure 3C). In addition, the expression of COLI and MMP1 was significantly increased in ADSCE2F1-/--Exo group compared with the other two groups (Figure 3D). Hydroxyproline (HYP) is one of the main components of collagen, and its content can reflect the degree of collagen synthesis.27 We found that the HYP content of the cell supernatant from ADSCE2F1-/--Exo group was significantly increased compared with that in the other two groups (Figure 3E). Simultaneously, ADSCE2F1-/--Exos and ADSCWT-Exos were applied to vascular endothelial cell C166. Wound healing assay and transwell assay demonstrated that ADSCE2F1-/--Exo treatment promoted the migration ability of C166 cells compared with the PBS and ADSCWT-Exo treatment (Figure 3F-G). Also, ADSCE2F1-/--Exos markedly enhanced the proliferation of C166 cells (Figure 3H). The tube formation assay revealed that ADSCE2F1-/--Exo-treated C166 cells presented increased tubule form compared with the other two groups (Figure 3I). These results suggested that ADSCE2F1-/--Exos promoted the migration and proliferation of C166 and NIH-3T3 cells and enhanced the collagen formation of NIH-3T3 cells and tube formation of C166 cells.
E2F1 Regulated miR-130b-5p Expression in ADSCs at the Transcriptional Level
MiRNAs are one of the most important substances in ADSC-Exos related to wound healing. MiRNA high-throughput sequencing was performed to screen the differently expressed miRNA between ADSCE2F1-/- and ADSCWT. The volcano plot presented 9 upregulated miRNAs and 3 downregulated miRNAs of ADSCE2F1-/- compared with ADSCWT (Figure 4A). The heat map showed the expression of these miRNAs in ADSC samples (Figure 4B). The PPI network maps showed the interaction between differentially expressed miRNAs and their target proteins (Figure S1). Through literature research, we found that miR-130b-5p played an important role in collagen formation and cell cycle regulation.28 We speculated that E2F1 knockout enhanced the ability of ADSC-Exos to promote wound healing by increasing the miR-130b-5p expression. qRT-PCR was performed to verify the results of miRNA high-throughput sequencing. Compared with ADSCWT, miR-130b-5p expression in ADSCE2F1-/- was increased (Figure 4C). Similarly, miR-130b-5p expression in ADSCs was upregulated after transfection of si-E2F1 (Figure 4D). Besides, miR-130b-5p expression was upregulated in ADSCE2F1-/--Exos compared with ADSCWT-Exos (Figure 4E). Analysis of ALGGEN database presented that there was a potential binding site of E2F1 on the promoter region of miR-130b-5p (Figure 4F). The ChIP assay verified the prediction of ALGGEN database, revealing that E2F1 was bound to the promoter region of miR-130b-5p (Figure 4G-H). These results indicated that E2F1 regulated miR-130b-5p transcription, and E2F1 knockout enhanced miR-130b-5p expression in ADSCs and ADSC-Exos.
ADSCE2F1-/--Exos Promoted Proliferation, Migration, and Collagen Formation of Fibroblasts via Transferring miR-130b-5p
Based on abovementioned results, we speculated that ADSCE2F1-/--Exos might affect fibroblasts via transferring miR-130b-5p. The expression of miR-130b-5p was upregulated in NIH-3T3 cells after ADSCE2F1-/--Exo treatment (Figure 5A). IF showed that PKH26-labelled ADSCE2F1-/--Exos were internalized into NIH-3T3 cells after 24 h co-culture (Figure 5B). The FISH assay suggested that the subcellular localization of miR-130b-5p was mainly located in the cytoplasm (Figure 5C). Following transfection with miR-130b-5p mimic and inhibitor in NIH-3T3 cells (Figure 5D), the results from wound healing, transwell, and EdU suggested that miR-130b-5p promoted the migration and proliferation of NIH-3T3 cells (Figure 5E–H). The HYP content assay suggested that miR-130b-5p promoted collagen formation in NIH-3T3 cells (Figure 5I).
MiR-130b-5p Promoted Proliferation, Migration, and Collagen Formation of Fibroblasts by Targeting TGFBR3
The miRBD, miRmap, miRWalk, and TargetScan databases were used to predict the target gene of miR-130b-5p (Figure 6A). TGFBR3 was one of the overlapped target genes and has been reported to regulate the function of fibroblasts.29 TargetScan predicted the binding sites of miR-130b-5p and TGFBR3 (Figure 6B). The dual-luciferase activity assay confirmed that TGFBR3 was the target gene of miR-130b-5p (Figure 6C). TGFBR3 expression was downregulated after transfection with miR-130b-5p mimic, and was upregulated after transfection with miR-130b-5p inhibitor (Figure 6D). Western bolt analysis showed that OE-TGFBR3 upregulated the expression of TGFBR3 (Figure 6E). Wound healing, transwell, and EdU proved that TGFBR3 overexpression reversed the effects of miR-130b-5p mimic on the NIH-3T3 cell proliferation, and migration (Figure 6F-I). The HYP content assay suggested that TGFBR3 overexpression decreased the collagen formation in NIH-3T3 cells (Figure 6J).
ADSCE2F1-/--Exos Activated TGF-β Signaling Pathway
To explore the potential mechanism of ADSCE2F1-/--Exos in affecting fibroblasts, the TGF-β signaling pathway changes in NIH-3T3 cells under different treatments were determined. Smad2 and Smad3 are the two main downstream regulators of TGF-β signaling pathway that regulate collagen formation. Our results revealed that ADSCE2F1-/--Exos upregulated the expression of p-SMAD2 and p-SMAD3, and downregulated the expression of TGFBR3 (Figure 7A and B). Besides, miR-130b-5p mimic and si-TGFBR3 also increased p-SMAD2 and p-SMAD3 expression and decreased TGFBR3 expression (Figure 7C and D). The IHC results showed that TGF-β expression was significantly increased in the wound tissue of the ADSCE2F1-/--Exo group at day 7 post-treatment, but with no significant difference with other groups at day 14 post-treatment (Figure 7E). These results suggested that ADSCE2F1-/--Exos delivered miR-130b-5p into fibroblasts, and ultimately activated the TGF-β signaling pathway in fibroblasts, by targeting and regulating TGFBR3 expression. Therefore, ADSCE2F1-/--Exos could activate TGF-β signaling pathway at the early stages.
Discussion
In this study, we determined that the local transplantation of ADSCE2F1-/--Exos in wound sites significantly enhanced wound regeneration by promoting angiogenesis and collagen deposition, and the wound healing effect of ADSCE2F1-/--Exos was better than that of ADSCWT-Exos. Furthermore, E2F1 loss upregulated miR-130b-5p transcription in ADSCs and the content of miR-130b-5p in ADSCE2F1-/--Exos. Intriguingly, ADSCE2F1-/--Exos delivered miR-130b-5p into fibroblasts and enhanced the function of fibroblasts via activating the miR-130b-5p/TGFBR3 signaling pathway.
Skin wound healing is a fundamental physiological process for maintaining skin integrity.30 Disorder in the wound healing process leads to bacterial colonization, refractory ulcers, and excessive scarring. Delayed wound healing is one of the most intractable problems for clinicians and brings economic burdens to patients.31 Therefore, shortening healing time and promoting tissue regeneration represent urgent clinical needs. Recently, ADSC-Exos have emerged as an attractive method in wound regeneration.30 Recently, ADSC-Exos had emerged as an attractive method in wound regeneration.32 ADSC-Exos possess diverse bioactive functions similar to ADSCs, with a lower risk of immune rejection and malignancy than ADSCs.33 For instance, ADSC-Exos facilitated skin repair by improving new blood vessel formation and collagen synthesis.34 Specific interpretation methods, such as drug preconditioning, control of culture conditions, and gene modification, could be utilized to improve the function of ADSCs-Exos. Hypoxia ADSC-derived exosomes improved wound healing via activating PI3K/Akt pathways.35 Exosomes derived from Nrf2-overexpressing ADSCs promoted diabetic wound healing via suppressing ROS and inflammatory cytokine secretion.36 Therefore, it is urgent to explore effective interpretation methods to improve the ability of ADSC to promote wound healing. E2F1 regulates the transcription of multiple miRNAs and is associated with tissue regeneration.37 Thus, we supposed that E2F1 knockout would reshape the paracrine expression profile of ADSC-Exos and improve the function of ADSC-Exos in promoting wound healing. Our data showed that E2F1 knockout could improve the ability of ADSC-Exos to promote wound healing. This suggested that E2F1 deletion can affect the secretion profile of ADSC-Exos, which motivated us to explore the specific targets and mechanisms by which E2F1 regulates ADSCs.
It is well known that transcription factor E2F1 is a crucial factor in regulating the cell cycle, oxidative stress, and cell death processes.38 Our previous research found that ADSCs from E2F1–/– mice promoted wound healing via enhancing VEGF and TGF-β1 paracrine.21 Besides, E2F1 knockout was protective in myocardial ischemia and skin injury.39,40 MiRNAs participate in various biological processes through post-transcriptional regulation, and miRNA expression can be regulated by transcription factors.41 Wang et al suggested that E2F1 knockout enhanced the miR-30b expression of cardiomyocytes to inhibit cardiomyocyte necrosis.42 Maixner et al demonstrated that E2F1 could upregulate the miR-206 and miR-210-5p expression of human visceral adipose tissue, which might contribute to extreme obesity.43 These researches proved that E2F1 could not only regulate the transcription of miRNAs, but also affect the paracrine expression profile of ADSCs. However, it remains ambiguous how E2F1 regulates the miRNA expression of ADSCs and ADSC-Exos. Thus, we hypothesize that E2F1 regulates the miRNA expression profile of ADSCs via transcriptional regulation, and then changes the miRNA contained in exosomes. Our research here illuminated that E2F1 knockout markedly up-regulated the miR-130b-5p expression in ADSCs via transcriptional regulation. Subsequently, miR-130b-5p could be secreted into recipient cells by exosomes.
MiRNAs are a class of short-stranded non-coding RNA types that can participate in a variety of pathophysiological processes by regulating the expression of target genes.44 Lu et al suggested that exosomal miR-486-5p derived from ADSCs promoted collagen synthesis and angiogenesis via targeting Sp5, thus accelerating wound healing.45 Besides, exosomal miR-125a-3p derived from ADSCs promoted the angiogenic capacity of HUVECs, and accordingly accelerated wound healing via inhibiting PTEN.46 Employing multi-omics sequencing and diverse analytical methods, more and more miRNAs are being characterized in terms of their expression abundance and species, and are expected to enhance the paracrine function of ADSC-Exos through miRNA overexpression or knockdown. Our research proved that miR-130b-5p derived from ADSCE2F1-/--Exos promoted the proliferation, migration, and collagen formation of fibroblasts. In previous studies, miR-130b-5p has been reported to protect against apoptosis and promote collagen formation. miR-130b-5p ameliorated neuronal damage induced by activated microglia via inhibiting the TLR4/NF-κB signaling pathway.47 Meanwhile, miR-130b-5p promoted COLI synthesis by activating the TGF-β pathway in mouse hepatic fibrosis models.28 Here, we emphasized that miR-130b-5p in ADSCE2F1-/--Exos was a protective factor for wound healing, and was closely related to TGF-β activation and promotion of collagen deposition.
TGFBR3 is one of the transmembrane receptors of the TGF-β superfamily.48 Downregulation of TGFBR3 induced by macrophage exosomes has been shown to promote migration, invasion, and angiogenesis of pancreatic cancer by activating the TGF-β signaling pathway.49 Exosomal miR-103a-3p from crohn’s creeping fat-derived ADSCs could induce TGFBR3 inhibition, and promote fibroblast activation in intestinal fibrosis via the TGF-β signaling pathway.29 Activation of the TGF-β signaling pathway is important in the early stages of wound healing and promotes the expression of genes associated with ECM formation to accelerate granulation tissue formation.50 In our study, ADSCE2F1-/--Exos activated the TGF-β signaling pathway in fibroblasts via miR-130b-5p/TGFBR3 axis and up-regulated TGF-β1 expression of wound tissue at the early stage of wound healing.
Nevertheless, there are still some limitations in our research. MiR-130b-5p may not be the only content in ADSCE2F1-/--Exos that accelerates wound healing. Other unexplored miRNAs may also possess such functions, improving wound healing by working together in a coordinated manner. Thus, further research is needed to explore the regulatory effect of E2F1 in other non-coding RNAs and cytokines in ADSC-Exos. Besides, as ADSCs were isolated from E2F1–/– C57BL/6 mice, E2F1 knockout in other organs may affect the function of ADSCs. Therefore, it is debatable whether it is better to select primary cells from gene knockout mice or to select primary cells followed by intervention using shRNA and other means. Additionally, there are also some challenges and limitations in ADSC-Exos themselves, including difficulties in large-scale production, standardization of isolation methods, and potential variations in therapeutic efficacy. Firstly, exosomes are mainly produced through cell culture and collection of conditioned media. Scaling up cell culture and achieving high yields of exosomes remain a challenge. Secondly, there is a lack of standardized isolation methods for exosomes. Currently, various methods including ultracentrifugation, filtration, and density gradient centrifugation are used for exosome isolation. However, differences in these methods may lead to variations in exosome purity and composition. Lastly, there are potential variations in therapeutic efficacy of exosomes. As exosomes are cell-derived vesicles, their composition and function can be influenced by factors such as cell source, culture conditions, and isolation methods. These factors may impact the consistency and predictability of their therapeutic effects. Therefore, further research is needed to achieve standardization and optimization of exosome production and separation methods to ensure the stability and reliability of their therapeutic efficacy.
Conclusion
In summary, we aimed to improve the therapeutic efficacy of ADSCs through genetic modification. Our findings provide evidence that E2F1 knockout improves the ability of ADSC to promote wound healing through the miR-130b-5p/TGFBR3 axis both in vivo and in vitro. Our results suggested that ADSCE2F1-/--Exos effectively enhanced wound healing in mice by promoting collagen formation and angiogenesis. E2F1 deficiency upregulated miR-130b-5p expression in ADSCE2F1-/--Exos. Besides, ADSCE2F1-/--Exos promoted the proliferation, migration, and collagen formation of fibroblasts via the miR-130b-5p/TGFBR3 axis. This study provides a novel method of gene-engineered ADSC-Exos for accelerating wound healing. However, it is important to note that miR-130b-5p may not be the only component within ADSCE2F1-/--Exos that contributes to the promotion of wound healing. Therefore, it would be valuable to investigate the regulatory effects of E2F1 on other cargo molecules present in ADSC-Exos. In addition, we mainly used mouse-derived ADSCs here, which is different from human ADSCs and has some differences in the application of clinical wound healing.
Abbreviations
ADSCs, Adipose-derived stem cells; ADSCE2F1-/--Exos, ADSCE2F1-/--derived exosomes; ADSCWT-Exos, ADSCWT-derived exosomes; BSA, bovine serum albumin; ChIP, chromatin immunoprecipitation; DLS, dynamic light scattering; ECM, extracellular matrix; FBS, fetal bovine serum; FISH, fluorescence in situ hybridization; H&E, hematoxylin and eosin; HYP, hydroxyproline; IF, immunofluorescence; IHC, immunohistochemistry; MSCs, mesenchymal stromal cells; miRNAs, microRNAs; siRNA, small interfering RNAs; SD, standard deviation; TEM, transmission electron microscope; EdU, 5-Ethynyl-20-Deoxyuridine.
Data Sharing Statement
All the datasets displayed in this study can be obtained in the manuscript. Further questions can be directed to the corresponding author Min Wu. E-mail address: [email protected].
Ethics Approval and Consent to Participate
All animal experiments were approved by the Animal Care Committee of Tongji Medical College (TJH-201909006) in September 30, 2019. The Laboratory Animal Guidelines for Ethical Review of Animal Welfare (GB/T 35892-2018) was followed to ensure the welfare of the laboratory animals.
Consent for Publication
All authors have provided their consent for publication.
Acknowledgments
Honghao Yu and Yiping Wu are co-first authors for this study.
Funding
This work was supported by The Science and Technology Project of Hubei Province (grant number: 2022CFB226), Research Foundation Project of Tongji Hospital (grant number:2022A16), China Guanghua Science and Technology Foundation (grant number: 2019JZXM001), and Wuhan Science and Technology Bureau (grant number: 2020020601012241).
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
1. Tripathi R, Knusel KD, Ezaldein HH, Honaker JS, Bordeaux JS, Scott JF. Incremental Health Care Expenditure of Chronic Cutaneous Ulcers in the United States. JAMA Dermatology. 2019;155(6):694–699. doi:10.1001/jamadermatol.2018.5942
2. Falanga V, Isseroff RR, Soulika AM, et al. Chronic wounds. Nat Rev Dis Prim. 2022;8:50. doi:10.1038/s41572-022-00377-3
3. Gurtner GC, Werner S, Barrandon Y, Longaker MT. Wound repair and regeneration. Nature. 2008;453:314–321. doi:10.1038/nature07039
4. Rodrigues M, Kosaric N, Bonham CA, Gurtner GC. Wound healing: a cellular perspective. Physiol Rev. 2019;99:665–706. doi:10.1152/physrev.00067.2017
5. Nosrati H, Heydari M, Tootiaei Z, Ganjbar S, Khodaei M. Delivery of antibacterial agents for wound healing applications using polysaccharide-based scaffolds. J Drug Deliv Sci Technol. 2023;84:104516. doi:10.1016/j.jddst.2023.104516
6. Al-Ghadban S, Bunnell BA. Adipose tissue-derived stem cells: immunomodulatory effects and therapeutic potential. Physiology. 2020;35:125–133. doi:10.1152/physiol.00021.2019
7. Chen S, He Z, Xu J. Application of adipose-derived stem cells in photoaging: basic science and literature review. Stem Cell Res Ther. 2020;11. doi:10.1186/s13287-020-01994-z
8. Cai Y, Li J, Jia C, He Y, Deng C. Therapeutic applications of adipose cell-free derivatives: a review. Stem Cell Res Ther. 2020;11:1–16. doi:10.1186/s13287-020-01831-3
9. Xiong M, Zhang Q, Hu W, et al. The novel mechanisms and applications of exosomes in dermatology and cutaneous medical aesthetics. Pharmacol Res. 2021;166:105490. doi:10.1016/j.phrs.2021.105490
10. Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367:eaau6977. doi:10.1126/science.aau6977
11. Zhou C, Zhang B, Yang Y, et al. Stem cell-derived exosomes: emerging therapeutic opportunities for wound healing. Stem Cell Res Ther. 2023;14:107. doi:10.1186/s13287-023-03345-0
12. Ragni E, Perucca Orfei C, De Luca P, et al. Interaction with hyaluronan matrix and miRNA cargo as contributors for in vitro potential of mesenchymal stem cell-derived extracellular vesicles in a model of human osteoarthritic synoviocytes. Stem Cell Res Ther. 2019;10. doi:10.1186/s13287-019-1215-z
13. Chen B, Cai J, Wei Y, et al. Exosomes Are Comparable to Source Adipose Stem Cells in Fat Graft Retention with Up-Regulating Early Inflammation and Angiogenesis. Plast Reconstr Surg. 2019;144:816E–827E. doi:10.1097/PRS.0000000000006175
14. Ma T, Fu B, Yang X, Xiao Y, Pan M. Adipose mesenchymal stem cell-derived exosomes promote cell proliferation, migration, and inhibit cell apoptosis via Wnt/β-catenin signaling in cutaneous wound healing. J Cell Biochem. 2019;120:10847–10854. doi:10.1002/jcb.28376
15. Chen S, Sun F, Qian H, Xu W, Jiang J. Preconditioning and Engineering Strategies for Improving the Efficacy of Mesenchymal Stem Cell-Derived Exosomes in Cell-Free Therapy. Stem Cells Int. 2022;2022:1–18. doi:10.1155/2022/1779346
16. Shi R, Jin Y, Zhao S, Yuan H, Shi J, Zhao H. Hypoxic ADSC-derived exosomes enhance wound healing in diabetic mice via delivery of circ-Snhg11 and induction of M2-like macrophage polarization. Biomed Pharmacother. 2022;153. doi:10.1016/j.biopha.2022.113463
17. Huang H, Xu Z, Qi Y, et al. Exosomes from SIRT1-Overexpressing ADSCs Restore Cardiac Function by Improving Angiogenic Function of EPCs. Mol Ther - Nucleic Acids. 2020;21:737–750. doi:10.1016/j.omtn.2020.07.007
18. Biswas AK, Johnson DG. Transcriptional and nontranscriptional functions of E2F1 in response to DNA damage. Cancer Res. 2012;72:13–17. doi:10.1158/0008-5472.CAN-11-2196
19. Wu M, Zhou J, Cheng M, et al. E2F1 suppresses cardiac neovascularization by down-regulating VEGF and PlGF expression. Cardiovasc Res. 2014;104:412–422. doi:10.1093/cvr/cvu222
20. Wang N, Wu Y, Zeng N, et al. E2F1 Hinders Skin Wound Healing by Repressing Vascular Endothelial Growth Factor (VEGF) Expression, Neovascularization, and Macrophage Recruitment. PLoS One. 2016;11:e0160411. doi:10.1371/journal.pone.0160411
21. Yi Z, Wu Y, Zhang Q, et al. E2F1- Deficient Adipose-Derived Stem Cells Improve Skin Wound Closure in Mice by Upregulating Paracrine Expression of VEGF and TGF-Β1. Int J Med. 2023. doi:10.1097/prs.0000000000010145
22. Meng Z, Zhou D, Gao Y, Zeng M, Wang W. miRNA delivery for skin wound healing ☆. Adv Drug Deliv Rev. 2018;129:308–318. doi:10.1016/j.addr.2017.12.011
23. Wang K, Zhou L-Y, Wang J-X, et al. E2F1-dependent miR-421 regulates mitochondrial fragmentation and myocardial infarction by targeting Pink1. Nat Commun. 2015;6:7619. doi:10.1038/ncomms8619
24. Qu Y, Zhang Q, Cai X, et al. Exosomes derived from miR-181-5p-modified adipose-derived mesenchymal stem cells prevent liver fibrosis via autophagy activation. J Cell Mol Med. 2017;21:2491–2502. doi:10.1111/jcmm.13170
25. Bieg D, Sypniewski D, Nowak E, Bednarek I. MiR-424-3p suppresses galectin-3 expression and sensitizes ovarian cancer cells to cisplatin. Arch Gynecol Obstet. 2019;299:1077–1087. doi:10.1007/s00404-018-4999-7
26. Li X, Liao J, Su X, et al. Human urine-derived stem cells protect against renal ischemia/reperfusion injury in a rat model via exosomal miR-146a-5p which targets IRAK1. Theranostics. 2020;10:9561–9578. doi:10.7150/thno.42153
27. Eid BG, Alhakamy NA, Fahmy UA, et al. Melittin and diclofenac synergistically promote wound healing in a pathway involving TGF-β1. Pharmacol Res. 2022;175:105993. doi:10.1016/j.phrs.2021.105993
28. Wang H, Wang Z, Wang Y, et al. miRNA-130b-5p promotes hepatic stellate cell activation and the development of liver fibrosis by suppressing SIRT4 expression. J Cell Mol Med. 2021;25:7381–7394. doi:10.1111/jcmm.16766
29. Qian W, Xu Y, Wen W, et al. Exosomal miR-103a-3p from Crohn’s Creeping Fat-Derived Adipose-Derived Stem Cells Contributes to Intestinal Fibrosis by Targeting TGFBR3 and Activating Fibroblasts. J Crohn’s Colitis. 2023;11:2662. doi:10.1093/ecco-jcc/jjad042
30. Martin P, Nunan R. Cellular and molecular mechanisms of repair in acute and chronic wound healing. Br J Dermatol. 2015;173:370–378. doi:10.1111/bjd.13954
31. Bosanquet DC, Harding KG. Wound healing: potential therapeutic options. Br J Dermatol. 2022;187:149–158. doi:10.1111/bjd.20772
32. An Y, Lin S, Tan X, et al. Exosomes from adipose‐derived stem cells and application to skin wound healing. Cell Prolif. 2021;54:1–12. doi:10.1111/cpr.12993
33. Xiong M, Zhang Q, Hu W, et al. Exosomes From Adipose-Derived Stem Cells: the Emerging Roles and Applications in Tissue Regeneration of Plastic and Cosmetic Surgery. Front Cell Dev Biol. 2020;8:1–17. doi:10.3389/fcell.2020.574223
34. Ren S, Chen J, Duscher D, et al. Microvesicles from human adipose stem cells promote wound healing by optimizing cellular functions via AKT and ERK signaling pathways. Stem Cell Res Ther. 2019;10:1–14. doi:10.1186/s13287-019-1152-x
35. Wang J, Wu H, Peng Y, et al. Hypoxia adipose stem cell-derived exosomes promote high-quality healing of diabetic wound involves activation of PI3K/Akt pathways. J Nanobiotechnology. 2021;19:1–13. doi:10.1186/s12951-021-00942-0
36. Li X, Xie X, Lian W, et al. Exosomes from adipose-derived stem cells overexpressing Nrf2 accelerate cutaneous wound healing by promoting vascularization in a diabetic foot ulcer rat model. Exp Mol Med. 2018;50. doi:10.1038/s12276-018-0058-5
37. Palm T, Hemmer K, Winter J, et al. A systemic transcriptome analysis reveals the regulation of neural stem cell maintenance by an E2F1-miRNA feedback loop. Nucleic Acids Res. 2013;41:3699–3712. doi:10.1093/nar/gkt070
38. Qin G, Kishore R, Dolan CM, et al. Cell cycle regulator E2F1 modulates angiogenesis via p53-dependent transcriptional control of VEGF. Proc Natl Acad Sci. 2006;103:11015–11020. doi:10.1073/pnas.0509533103
39. Xu S, Tao J, Yang L, et al. E2F1 Suppresses Oxidative Metabolism and Endothelial Differentiation of Bone Marrow Progenitor Cells. Circ Res. 2018;122:701–711. doi:10.1161/CIRCRESAHA.117.311814
40. Xiao H, Wu YP, Yang CC, et al. Knockout of E2F1 enhances the polarization of M2 phenotype macrophages to accelerate the wound healing process. Kaohsiung J Med Sci. 2020;36:692–698. doi:10.1002/kjm2.12222
41. Zhang H-M, Kuang S, Xiong X, Gao T, Liu C, Guo A-Y. Transcription factor and microRNA co-regulatory loops: important regulatory motifs in biological processes and diseases. Brief Bioinform. 2015;16:45–58. doi:10.1093/bib/bbt085
42. Wang K, An T, Zhou L-Y, et al. E2F1-regulated miR-30b suppresses Cyclophilin D and protects heart from ischemia/reperfusion injury and necrotic cell death. Cell Death Differ. 2015;22:743–754. doi:10.1038/cdd.2014.165
43. Maixner N, Haim Y, Blüher M, et al. Visceral Adipose Tissue E2F1-miRNA206/210 Pathway Associates with Type 2 Diabetes in Humans with Extreme Obesity. Cells. 2022;11:3046. doi:10.3390/cells11193046
44. Bian D, Wu Y, Song G, Azizi R, Zamani A. The application of mesenchymal stromal cells (MSCs) and their derivative exosome in skin wound healing: a comprehensive review. Stem Cell Res Ther. 2022;13:24. doi:10.1186/s13287-021-02697-9
45. Lu Y, Wen H, Huang J, et al. Extracellular vesicle‐enclosed miR‐486‐5p mediates wound healing with adipose‐derived stem cells by promoting angiogenesis. J Cell Mol Med. 2020;24:9590–9604. doi:10.1111/jcmm.15387
46. Pi L, Yang L, Fang B-R, Meng -X-X, Qian L. Exosomal microRNA-125a-3p from human adipose-derived mesenchymal stem cells promotes angiogenesis of wound healing through inhibiting PTEN. Mol Cell Biochem. 2022;477:115–127. doi:10.1007/s11010-021-04251-w
47. Wang D, Zhao S, Pan J, et al. Ginsenoside Rb1 attenuates microglia activation to improve spinal cord injury via microRNA-130b-5p/TLR4/NF-κB axis. J Cell Physiol. 2021;236:2144–2155. doi:10.1002/jcp.30001
48. Vander Ark A, Cao J, Li X. TGF-β receptors: in and beyond TGF-β signaling. Cell Signal. 2018;52:112–120. doi:10.1016/j.cellsig.2018.09.002
49. Yin Z, Ma T, Huang B, et al. Macrophage-derived exosomal microRNA-501-3p promotes progression of pancreatic ductal adenocarcinoma through the TGFBR3-mediated TGF-β signaling pathway. J Exp Clin Cancer Res. 2019;38:310. doi:10.1186/s13046-019-1313-x
50. Morikawa M, Derynck R, Miyazono K. TGF-β and the TGF-β Family: context-Dependent Roles in Cell and Tissue Physiology. Cold Spring Harb Perspect Biol. 2016;8:a021873. doi:10.1101/cshperspect.a021873
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.