Back to Journals » International Journal of Nanomedicine » Volume 19
Exosomes as Powerful Biomarkers in Cancer: Recent Advances in Isolation and Detection Techniques
Authors Zhang Q, Wang H, Liu Q, Zeng N, Fu G, Qiu Y, Yang Y, Yuan H, Wang W, Li B
Received 18 December 2023
Accepted for publication 13 February 2024
Published 26 February 2024 Volume 2024:19 Pages 1923—1949
DOI https://doi.org/10.2147/IJN.S453545
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor R.D.K. Misra
Qiongdan Zhang,1,2,* Huizhen Wang,1,2,* Qingyi Liu,1,2 Ni Zeng,1,2 Gang Fu,1,2 Yixing Qiu,1,2 Yupei Yang,1,2 Hanwen Yuan,1,2 Wei Wang,1,2 Bin Li1,2
1TCM and Ethnomedicine Innovation & Development International Laboratory, School of Pharmacy, Hunan University of Chinese Medicine, Changsha, People’s Republic of China; 2School of Pharmacy, Hunan University of Chinese Medicine, Changsha, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Wei Wang; Bin Li, Tel +86-136-5743-8606 ; +86-158-7410-0917, Email [email protected]; [email protected]
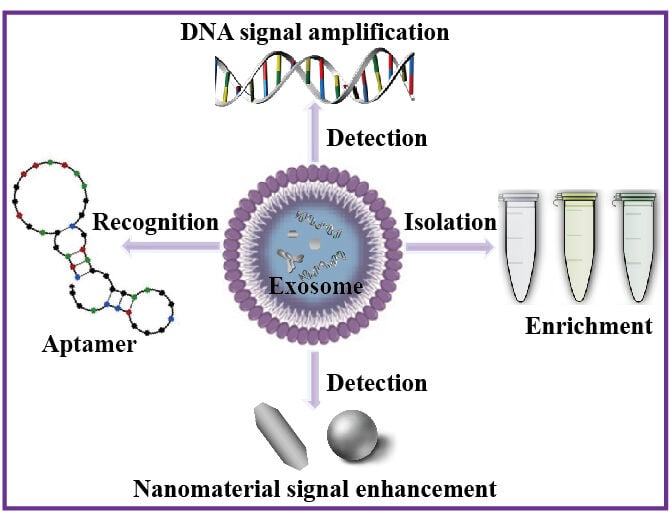
Abstract: Exosomes, small extracellular vesicles derived from cells, are known to carry important bioactive molecules such as proteins, nucleic acids, and lipids. These bioactive components play crucial roles in cell signaling, immune response, and tumor metastasis, making exosomes potential diagnostic biomarkers for various diseases. However, current methods for detecting tumor exosomes face scientific challenges including low sensitivity, poor specificity, complicated procedures, and high costs. It is essential to surmount these obstacles to enhance the precision and dependability of diagnostics that rely on exosomes. Merging DNA signal amplification techniques with the signal boosting capabilities of nanomaterials presents an encouraging strategy to overcome these constraints and improve exosome detection. This article highlights the use of DNA signal amplification technology and nanomaterials’ signal enhancement effect to improve the detection of exosomes. This review seeks to offer valuable perspectives for the enhancement of amplification methods applied in practical cancer diagnosis and prognosis by providing an overview of how these novel technologies are utilized in exosome-based diagnostic procedures.
Keywords: tumor, exosomes, nanomaterials, DNA signal amplification
Graphical Abstract:
Introduction
Exosomes, which are minute vesicles enclosed by a phospholipid bilayer membrane, are secreted by various cells in the human body.1–7 These vesicles fall under the classification of extracellular microvesicles (EVs) and typically measure between 40–160 nm in diameter.8–10 They play a crucial role in transmitting bioactive molecules (such as nucleic acid molecules, proteins, lipids) to specific recipient cells, enabling intercellular communication (Figure 1).11
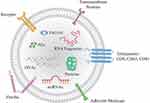 |
Figure 1 Structure and contents of exosome. Note: Reprinted from Chang KL, Sun P, Dong X, et al. Aptamers as recognition elements for electrochemical detection of exosomes. Chem Res Chin Univ. 2022;38(4):879–885.11 |
Nucleic acid molecules, such as RNA, including primarily microRNAs (miRNA) and messenger RNA (mRNA), as well as piRNA and tRNA discovered later on.12–14 In addition, exosomes are abundant in protein molecules, such as transmembrane proteins (CD9, CD63, and CD81, among others) that play a critical role in cell adhesion.15 Heat shock protein (HSP90) involved in protein folding processes can also be found in exosomes, as can proteins related to endosomal system transport (TSG101, Alix, and ESCRT family proteins).16,17 Notably, CD63, Alix, and TSG101 are proteins specifically expressed on the surface of exosomes, making them valuable biomarkers for identifying exosomes. Moreover, the exosome membrane surface is rich in sphingomyelin, cholesterol, and ceramide raft lipids.18
In the past few years, exosomes have gained increasing attention in tumor-related research due to their advantages, including high content, broad source, easy accessibility, structural stability, and outstanding biological characteristics. They are particularly valuable as an emerging non-invasive marker for early tumor diagnosis and therapeutic monitoring. For example, Liang’s group illustrated that the miRNA miR-25-3p, found within exosomes, facilitates metastasis in colorectal cancer (CRC) by enhancing vascular permeability and angiogenesis.19 In addition, He’s group revealed a significant increase in the expression of miR-21-5p in exosomes found in the bloodstream of colorectal cancer (CRC) patients compared to healthy individuals.20 The transmission of miR-21-5p via exosomes from CRC cells to endothelial cells results in an increased abundance of miR-21-5p within the recipient cells. Subsequently, miR-21-5p suppresses the activity of Krev interaction trapped protein 1 (KRIT1) in HUVECs, leading to the activation of the β-catenin signaling pathway and elevated expression of VEGFa and Ccnd1 downstream targets. As a result, this process ultimately stimulates angiogenesis and enhances vascular permeability in CRC. Similarly, Liang et al conducted a study showing that the down-regulation of miR-21 resulted in cell cycle arrest, reduced proliferation of HCT-1165FR, and increased apoptosis in a colorectal cancer cell line.21 Furthermore, in the study conducted by Ma’s group, it was found that miR-3157-3p expression levels in circulating exosomes were significantly higher in patients with metastatic non-small cell lung cancer (NSCLC) compared to those with non-metastatic NSCLC.22 This specific miR-3157-3p present in exosomes plays a critical role in the formation of pre-metastatic niches before tumor metastasis and holds promise as a potential blood-based biomarker for NSCLC metastasis. Through its targeting of TIMP/KLF2, exosome miR-3157-3p regulates the expression of VEGF/MMP2/MMP9 and occludin in endothelial cells, thereby facilitating angiogenesis and increasing vascular permeability. Meanwhile, Sandfeld-Paulsen’s group reported that approximately 72% of exosomes purified from non-small cell lung cancer (NSCLC) biopsies contained epidermal growth factor receptor (EGFR) proteins.23 Similarly, Huang’s group reported that approximately 80% of the exosomes isolated from lung cancer biopsy samples were found to contain EGFR.24 Yao group showed that exosomal miR-27a-3p has been shown to contribute to immune evasion in breast cancer by enhancing the expression of PD-L1 through the MAGI2/PTEN/PI3K pathway.25 These discoveries underscore the promise of exosomes as biomarkers for disease control and prevention, given their capacity to mirror the metabolic condition of their source cells and their role in essential pathogenic processes. As a result, precise identification and examination of exosomes specific to diseases are imperative for diverse uses in disease study and diagnostic procedures.
Recently, a multitude of detection methods have been established for the analysis of exosomes and their contents, serving both research and clinical objectives. Two traditionally used techniques include nanoparticle tracking analysis (NTA) and enzyme-linked immunosorbent assay (ELISA).26–29 NTA is capable of determining particle sizes ranging from 30 to 1000 nm using laser light scattering microscopy and a charge-coupled device (CCD) camera to visualize and record nanoparticles in solution. It is important to note that NTA has limitations in terms of repeatability and sensitivity when used for exosome analysis. On the other hand, ELISA involves purchasing a commercial ELISA kit for analyzing and detecting particles. Nevertheless, ELISA has some disadvantages, including its high cost, lengthy assay times, labor-intensive sample handling, and multiple steps involving reagent addition, washing, and incubation.30 Current detection methods often suffer from deficiencies like low sensitivity, low specificity, and cumbersome operation.31–33 Thus, there exists a need to create exosome detection techniques that are user-friendly while also being highly sensitive and specific. The objective of this review is to provide a comprehensive summary of different strategies employed to enhance the sensitivity and specificity in exosome detection, utilizing aptamer-based specific recognition, nanomaterials signal enhancement (Quantum dots, Au nanoparticles, Graphene oxide nanosheets, etc) and DNA-based signal amplification techniques (Catalytic hairpin assembly (CHA), Hybrid chain reaction (HCR), and Rolling cycle amplification (RCA), etc).
Isolation Techniques for Exosomes
Exosomes are commonly found in a range of biofluids, such as blood, urine, sweat, and others.34–37 However, isolating and enriching exosomes from these complex biofluids can be challenging due to the presence of numerous biomacromolecules and protein aggregates.38–41 The presence of these constituents, particularly in clinical samples, can impede the precise detection and analysis of exosomes. To overcome this challenge, various techniques based on size, density, and surface proteins have been able to be devised for the isolation and enrichment of exosomes from complex biofluids.42 Differential ultracentrifugation and density gradient centrifugation are two commonly employed methods for isolating exosomes from complex biofluids.43–46 Additionally, there are commercially available kits and reagents specifically designed for exosome isolation from biofluids. These kits often utilize proprietary methods such as precipitation, immunoaffinity capture, or size-exclusion chromatography is employed to selectively isolate exosomes while minimizing interference from other components present in the biofluids.47–51 Table 1 provides a comprehensive overview of various isolation methods for exosomes.
 |
Table 1 An Overview on Different Isolation Methods for Exosome |
Ultracentrifugation
Ultracentrifugation is a method of separating of particles based on their size and density. It can be categorized into two primary types: differential ultracentrifugation and density gradient ultracentrifugation.56,57
In differential ultracentrifugation, the separation of particles is achieved through the application of varying centrifugal forces and durations. Initially, the procedure employs relatively low centrifugal forces (in excess of 10,000 g) to facilitate the segregation of larger entities, including cells, cellular remnants, and sizable extracellular vesicles (EVs).58 Subsequently, strong centrifugal forces (100,000–200,000 g) and longer centrifugal times (<70 min) are employed to collect exosomes,52 Following the centrifugation process, a pellet enriched with exosomes is collected. This pellet is then thoroughly washed with phosphate-buffered saline (PBS) to strip away any proteins that are still present. During the washing step, the purified exosomes are obtained through centrifugal forces at 100,000 g for 30 min, and subsequently are redissolve at PBS and stored at a low temperature of −80 °C for preservation. (Figure 2A).
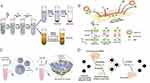 |
Figure 2 (A) Exosome isolation procedures based on ultracentrifugation. Reprinted from Bu HC, He DG, He XX, et al. Exosomes: isolation, analysis, and applications in cancer detection and therapy. Chembiochem. 2019;20(4):451–461. © 2019 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim.52 (B) Size-dependent microfluidics for exosome isolation. Reprinted from Liu C, Guo JY, Tian F, et al. Field-free isolation of exosomes from extracellular vesicles by microfluidic viscoelastic flows. ACS Nano. 2017;11(7):6968–6976. Copyright: American Chemical Society, 2017.54 (C) PEG precipitation for exosome isolation. Reprinted from Weng YJ, Sui ZG, Shan YC, et al. Effective isolation of exosomes with polyethylene glycol from cell culture supernatant for in-depth proteome profiling. Analyst. 2016;141(15):4640–4646. Copyright: Royal Society of Chemistry, 2016.55 (D) Immunoaffinity for exosome isolation. Reprinted from Chen JC, Li PL, Zhang TY, et al. Review on strategies and technologies for exosome isolation and purification. Front Bioeng Biotechnol. 2022;9:811971. Creative Commons.56 |
Density gradient ultracentrifugation is a more stringent method for separating exosomes. It relies on the buoyant density of exosomes in a sucrose density gradient. The buoyancy density of exosomes in sucrose aqueous solutions is usually between 1.10 to 1.20 g mL−1.52 The characteristic of these particles enables their separation via sucrose gradient centrifugation. This technique entails the creation of a sucrose density gradient within a centrifuge tube, with the gradient’s density progressively diminishing from the bottom upwards. Similar to differential ultracentrifugation, samples are initially processed in the same manner before being subjected to density gradient ultracentrifugation. Following this step, various extracellular vesicles (EVs) segregate into discrete bands along the gradient, sorted according to their respective densities. (Figure 2A). Density gradient ultracentrifugation is favored over differential ultracentrifugation due to its superior ability to maintain the biological integrity and characteristics of exosomes. As a result, this method is extensively employed for the enrichment of exosomes.59,60 However, it is noteworthy that ultracentrifugation, encompassing both differential and density gradient methods, has certain limitations: (1) costly equipment, extended operation durations, and substantial sample volumes are necessary; (2) the intense centrifugal forces applied during ultracentrifugation have the potential to compromise the structural integrity of exosomes, which can impede subsequent analytical procedures.61
Size-Based Isolation Techniques
Size-based techniques can be employed to isolate exosomes, which have smaller diameters compared to microvesicles and apoptotic bodies. These techniques provide advantages such as quicker isolation times and the absence of specialized equipment. Currently, two primary methods for size-based isolation are sequential filtration and size-dependent microfluidics.62
Sequential Filtration: Sequential filtration operates similarly to traditional filtration techniques. It relies on the molecular weight and size of different particles for their isolation.53 The process involves using membrane filters with varying exclusion limits based on size or molecular weight. First, cells, cell debris, and large extracellular vesicles (EVs) are eliminated through prefiltration using a 0.22 μm membrane filter. Then, free proteins are separated out using a 500 kDa molecular weight dialysis bag.53 Finally, the concentrated samples are filtered using a 100 nm membrane filter. Due to the gentle manipulation forces involved, sequentially filtered exosomes exhibit high purity.
Size-dependent Microfluidics: This method is additionally utilized for the separation of exosomes. The viscoelastic microfluidic apparatus comprises inlet I, inlet II, and inlet III. (Figure 2B).54 The sample fluid is introduced into the microchannel via inlet I, and simultaneously, a dilute solution of poly oxyethylene (PEO) is injected through inlet II. Within the microchannel, the flow of the sample fluid propels the extracellular vesicles (EVs) forward. Ultimately, larger EVs gravitate towards the microchannel’s centerline, while exosomes are directed towards the sides of the channel.63–65
Size-dependent microfluidics offer several advantages for exosome isolation. By reducing the device size, sample volume requirements are decreased, throughput is increase, and sensitivity is improved. These microfluidic platforms use capture agents within the device and surface biomarkers on exosomes to achieve highly specific and pure isolation. The microfluidic system is highly promising for exosome analysis and future applications, owing to its versatility and label-free capability for exosome separation by size.
Coprecipitation
Polymers like polyethylene glycol (PEG) are utilized to coprecipitate hydrophobic proteins and lipid molecules. This technique has proven effective for the isolation and purification of viruses. Given that exosomes exhibit biophysical characteristics akin to those of viruses, the PEG-based approach can be adapted for exosome isolation. (Figure 2C).55 To isolate exosomes using this method, the sample (such as culture supernatant or other biofluids) is first processed to remove cells and cellular debris. Subsequently, it is incubated with a PEG solution at 4 °C for 14 hours. Following centrifugation at low speeds, the sedimented extracellular vesicle (EV) products are located at the base of the centrifuge tube. This method is favored by numerous researchers for exosome separation because it is simple to execute and bypasses the extensive steps involved in ultracentrifugation. While coprecipitation offers an uncomplicated approach to isolate exosomes in just a few stages, it is not without its drawbacks. The relatively low throughput and potential contamination with chemicals can restrict its application in certain contexts. However, when these limitations are not a concern, the PEG-based method can be an effective and efficient approach for isolating exosomes.
Therefore, many researchers prefer the method of exosome separation that bypasses time-consuming ultracentrifugation due to its convenience. The coprecipitation technique offers a straightforward isolation process, but it has limitations in terms of its relatively low throughput and potential chemical contamination, which can restrict its broader application.
Immunoaffinity Enrichment
Immunoaffinity Based Capture (IAC) is a method that leverages the binding affinity between proteins present on exosome membranes and their respective protein receptors. This enables the specific isolation of exosomes from biological fluids. (Figure 2D).56 One common method that employs IAC is ELISA, which captures and quantifies exosomes using specific exosome markers such as CD63, CD326, and Tim-4 binding phosphatidylserine.66
IAC can also be utilized for additional purification of exosomes that were initially isolated using non-specific techniques based on density and size. When isolating tumor-specific exosomes, which are typically present in very low quantities, IAC presents a challenging task. Nonetheless, studies have shown that the IAC-based method employing mAb 763.74, which specifically targets the CSPG4 epitope expressed by melanoma cells, successfully separated and isolated melanoma-specific exosomes with an efficiency of approximately 95%.67 This method proved to be effective for liquid biopsy, offering potential applications in cancer diagnostics. These examples highlight the utility of IAC in specifically capturing and isolating exosomes of interest, including tumor-specific exosomes, enabling their potential application in liquid biopsies and disease diagnostics. Similarly, an anti-CD34 antibody-based IAC method successfully isolated acute myeloid leukemia (AML)-specific exosomes from cell culture supernatants. The CD34 microbeads employed in this approach demonstrated high efficiency, as only 10 µL aliquots were capable of capturing all the exosomes present in 100–1000 µL of AML suspension.68 These examples highlight the utility of IAC in specifically capturing and isolating exosomes of interest, including tumor-specific exosomes, enabling their potential application in liquid biopsies and disease diagnostics.
Thanks to the strong binding affinity between proteins and protein receptors, IAC offers several advantages in exosome separation, including high purity and efficiency. However, the use of antibodies, magnetic beads, and microbeads in this process has introduced expensive drawbacks to the method.
Overall, to select the most suitable approach for exosome isolation from complex biofluids, it is crucial to consider the specific requirements of the downstream application. By doing so, you can ensure that the chosen method will meet the necessary criteria for downstream analyses. It is recommended to thoroughly evaluate the efficiency, yield, purity, and compatibility of each method before making a decision.
Detection of Exosomes
Indeed, the isolation and analysis of exosomes have seen significant advancements in recent years, with a focus on achieving both high sensitivity and specificity. Numerous techniques and methods have been developed to tackle the challenges associated with exosome collection and detection, particularly low concentrations and complex sample backgrounds. One promising approach involves the use of aptamers for exosome capture and signal amplification.
Aptamer Based Exosomes Detection
Aptamers are artificially synthetic single-stranded DNAs or RNAs that consist of 25–80 bases and form unique secondary and tertiary structures upon binding with targets.69 They are commonly referred to as “chemical antibodies” due to their exceptional ability to recognize targets with high affinity and specificity, surpassing the advantages of traditional antibodies.69 Aptamers can be chemically produced, easily stored, and exhibit favourable attributes such as good stability, low immunogenicity, and toxicity levels. Aptamers can be confidently used in various DNA-based amplification reactions without compromising target binding.69 These exceptional quality makes aptamers indispensable for detecting and identifying exosomes, thereby contributing significantly to diagnostic applications. Recently, both single aptamer and double aptamer approaches have gained widespread use in exosome detection. Table 2 presents a comprehensive summary of aptamer-based exosome detection.
 |
Table 2 An Overview on Aptamer-Based Exosome Detection |
Single aptamer: The use of single nucleic acid aptamers in fluorescence and electrochemical sensors for exosome detection has gained significant popularity due to their remarkable binding affinity and selectivity towards the target. For instance, Kuang’s group proposed a colorimetric aptasensor for sensitive exosome detection based on a hemin/EpCAM aptamer DNAzyme (Figure 3A).70 The EpCAM aptamer, which is enriched in guanine, adopts a G4 structure and generates a hemin/G4 complex DNAzyme without the requirement for any specific sequence. A color reaction with TMB and H2O2 is catalyzed by this DNAzyme. Upon addition of exosomes, the EpCAM aptamer binds specifically to them, resulting in destruction of the DNAzyme structure and the loss of catalytic activity during color reaction. The absorbance demonstrated an inverse correlation with the concentration of exosomes. This method displays a favorable linear range from 106–108 particles/mL, with a detection limit (LOD) of 3.94×105 particles/mL. The biosensing strategy is highly cost-effective and user-friendly, as it does not require any alterations to the original aptamer sequence or complex signal amplification strategies. Moreover, it can rapidly detect multiple cancer exosomes within an hour, making it a significantly more convenient alternative to existing detection technologies. To further enhance the detection sensitivity, Li’s team constructed a separable visual aptasensor using double spherical nucleic acids (DSNAs) and terminal deoxynucleotidyl transferase (TdT) for the sensitive detection of exosomes (Figure 3B).71 In this approach, the initial step involves using a CD63 aptamer-modified magnetic beads (MNPs-AptCD63) as the first spherical nucleic acids to efficiently capture exosomes. This magnetic separation method prevents the aggregation of gold nanoparticles (GNPs) caused by complex components in the reaction solution. The second spherical nucleic acids comprise numerous nucleolin aptamer-modified GNPs (GNPs-Apt nucleolin) for the specific identification of exosomes. These nucleolin aptamers then generate long polyT sequences via TdT extension and induce the release of polyA from the GNPs-polyA (polyA-attached gold nanoparticles) by hybridization. This leads to salt-induced GNPs aggregation, leading to a color change that enables visual analysis of exosomes. This strategy achieves an impressively low LOD, as low as 45 particles/μL. The colorimetric sensor can confidently distinguish between clinical serum samples from healthy individuals and those with leukemia. Its primary advantage is the ability to detect the target through observing the color change in gold nanomaterials using naked eye, without requiring expensive equipment. This discovery confidently offers a promising avenue for future development of exosome-based liquid biopsies. To achieve high sensitivity in complex samples and reduce the interference caused by serum proteins, An’s research group has designed an electrochemical aptasensor for the ultrasensitive detection of tumor exosomes, which is based on click chemistry and the DNA hybridization chain reaction (HCR) for signal amplification (Figure 3C).72 In order to capture exosomes, the CD63 aptamer was initially immobilized on a glassy carbon electrode. Functionalized lipid electrophiles, specifically 4-oxo-2-nonenal alkyne (alkynyl-4-ONE) molecules, were then conjugated to the exosomes by reactions between amino and aldehyde groups. Click chemistry facilitated the attachment of azide-labeled DNA probe as exosomes anchor. The signal was amplified by hybridization chain reaction (HCR), and the connected horseradish peroxidase (HRP) molecules catalyzed the reaction of o-phenylenediamine (OPD) with H2O2. The concentration of exosomes was quantified by monitoring the electrochemical reduction current of 2.3-diaminophenazine (DAP). This proposed method enables the sensitive detection of exosomes within the range of 1.12×102 to 1.12×108 particles/µL, with a LOD of 96 particles/µL. The technique employed aptamers and click chemistry to achieve highly accurate detection of exosomes. The click chemistry reaction effectively reduced interference from serum proteins, providing strong evidence for the substantial potential of quantifying tumor exosomes in clinical diagnostics.
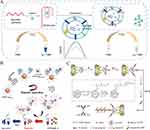 |
Figure 3 (A) Schematic representation of the hemin/EpCAM aptamer aptasensor for exosome detection. Reprinted from Kuang JJ, Fu ZB, Sun XZ, et al. A colorimetric aptasensor based on a hemin/EpCAM aptamer DNAzyme for sensitive exosome detection. Analyst. 2022;147(22):5054–5061. Copyright: Royal Society of Chemistry, 2022.70 (B) Schematic illustration of the presented colorimetric aptasensor for exosome detection. Reprinted from Li C, Wang HY, Wei R, et al. An excellent colorimetric aptasensor integrating multifunctional SNAs and TdT-induced dual signal amplification for rapid sensitive detection of exosomes. Sens Actuators B Chem. 2023;380:133361.71 (C) Schematic illustration of the proposed aptasensor for exosome detection based on click chemistry and HCR for signal amplification. Reprinted from An Y, Jin TY, Zhu YY, et al. An ultrasensitive electrochemical aptasensor for the determination of tumor exosomes based on click chemistry. Biosens Bioelectron. 2019;142:111503.72 |
Dual aptamer: The utilization of dual aptamers in sensor methods for exosome detection has gained significant attention due to their many advantages over single aptamers, such as enhanced sensitivity, improved target recognition specificity, and increased accuracy. For example, Zhu’s group proposed a dual labeling technique that utilizes protein-specific aptamer labeling and metabolic glycan labeling to visualize the glycosylation of specific proteins on exosomes. (Figure 4A).73 Within this method, initially, the exosomal sialic acids and PD-L1 proteins underwent a sequential labeling process: metabolic glycan labeling was performed on the exosomes using cellular MGL during exosome secretion, succeeded by the copper-free bioorthogonal conjugation of alkynyl DBCO-Cy5, and culminating with the precise targeting of PD-L1 proteins by a Cy3-tagged PD-L1 aptamer. Following this, the glycosylation of exosomal PD-L1 (exoPD-L1) was delineated in situ through an intramolecular FRET signal occurring between Cy3 and Cy5 on the same protein within the exosome. This method, both efficient and non-destructive, serves as a valuable technique for in situ analysis of the presence and functionality of glycosylation on specific exosomal proteins. In terms of the detection of exosomal proteins, You’s group developed a novel electrochemical biosensor that utilizes a hierarchical Au nanoarray-modified 2D Ti2CTx MXene membranes to detect exosomes. (Figure 4B).74 The Ti2CTx MXene membrane with a substantial specific surface area, combined with hierarchical gold nanoarrays known for their outstanding conductivity, enhances electrocatalytic properties and increased the number of active sites available for aptamer immobilization. In this technique, the composite membrane, functionalized with an EpCAM-targeting aptamer, was able to selectively capture exosomes. The captured exosomes were then used to anchor a CD63-specific aptamer, thereby enhancing the biosensor’s detection sensitivity and precision. The biosensor demonstrated exceptional sensitivity and dependable functionality in detecting exosomes, attaining a low LOD of 58 particles/μL within a linear detection range from 1×102 to 1×107 particles/μL. The sensor makes use of the modification of two distinct aptamer-target proteins. This modification facilitates accurate identification and quantification of cancer-specific exosomes while showcasing a strong capability to resist interference. Consequently, it enables effective quantification of exosomes in complex serum matrices, which greatly benefits the diagnostic analysis of clinical samples. In order to improve the accuracy of detection, Yu et al introduced a novel approach based on dual aptamer recognition and entropy-driven amplification for precise analysis of exosomes. (Figure 4C).75 There are two primary steps involved: 1) dual aptamer-based recognition of exosomes, and 2) entropy-driven catalytic system-based signal recycling. In the initial recognition process, the SMBs-S1 probe (containing CD63 aptamer) and S2-S4 probe complex (containing EpCAM aptamer) were employed for cooperative identification of exosomes. Subsequently, the S2–S4 probe complex disengaged the S4 probe via a chain displacement reaction with S5. This liberation of the S4 probe initiated an entropy-driven catalytic reaction that amplified the signal, thereby increasing the sensitivity of the method. Remarkably, the dual recognition of CD63 and EpCAM proteins endowed the method with outstanding specificity and stability, even amidst the presence of unbound CD63 and/or EpCAM proteins. The method displayed an extensive detection range spanning from 1×102 to 1×106 particles/μL, achieving a strong correlation coefficient (R2) of 0.9973. By utilizing the signal recycling-based entropy-driven catalytic method, this biosensor achieved exceptionally efficient signal amplification without relying on enzymes. This approach has enormous potential for screening diverse exosomes and serves as a versatile platform. The biosensor’s high accuracy and effectiveness also enable it to detect other biologically relevant molecules, holding tremendous promise for the early diagnosis and treatment of diseases.
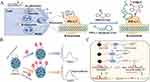 |
Figure 4 (A) Schematic diagram of exosomal aptamer-MGL strategy for characterizing exoPD-L1 glycosylation. Reprinted from Zhu L, Xu YF, We XY, et al. Coupling aptamer-based protein tagging with metabolic glycan labeling for in situ visualization and biological function study of exosomal protein-specific glycosylation. Angew Chem Int Ed Engl. 2021;60(33):18111–18115.73 (B) Schematic illustration of the construction of dual aptamer and Au–Ti2CTx membrane-based biosensor for exosome detection. Reprinted from You QN, Zhuang LL, Chang ZM, et al. Hierarchical Au nanoarrays functionalized 2D Ti2CTx MXene membranes for the detection of exosomes isolated from human lung carcinoma cells. Biosens Bioelectron. 2022;216:114647.74 (C) Schematic mechanism of dual aptamer recognition and entropy-driven catalytic system based for exosomes detection. Reprinted from Yu CX, Li L, Liu LH, et al. Research of exosome in bone metastasis through dual aptamer recognition based entropy-driven amplification. Anal Biochem. 2022;636:114433.75 |
In a word, a wide array of aptamers has been chosen and employed as innovative biological molecules for detecting membrane proteins associated with exosomes, primarily attributed to the strong affinity between the aptamers and their respective exosomes. Currently, sensors incorporating multiple aptamers have exhibited exceptional sensitivity and specificity in analyzing exosomes. In the future, integrating these sensors with advanced DNA computation technology will enable the acquisition of more comprehensive information about exosomes, thereby presenting abundant opportunities for clinical applications.
Nanomaterial Based Exosomes Detection
In recent times, nanomaterials have garnered significant attention owing to their small size and distinct optical and electromagnetic properties. These characteristics enable nanomaterials to interact with biological components at the microscopic level. This has led to their excellent biocompatibility and bioactivity, making them attractive for use in various biomedical applications. One area where nanomaterials have shown great promise is in the labeling, enrichment, counting, and detection of exosomes. Nanomaterials can also provide increased sensitivity and selectivity in exosome detection, thanks to advancements in imaging, spectroscopy, electromagnetic, and equipment technologies.76 Various types of nanomaterials have been explored for their use in exosome detection, such as quantum dots, Au nanoparticles, GONs nanomaterial, magnetic nanoparticles, etc. These nanomaterials can be engineered to emit or absorb specific wavelengths of light, allowing for their detection using spectroscopic techniques. They can also be functionalized with targeting molecules, enabling specific binding to exosomes of interest. Table 3 presents a detailed summary of nanomaterial-based exosome detection methods.
 |
Table 3 An Overview on Nanomaterial-Based Exosome Detection |
Quantum Dots Material Based Exosome Detection
Quantum dots refer to quasi-zero-dimensional nanomaterials with a particle size between 2–20 nm, consisting of a small number of atoms. Because quantum dots have unique, excellent optical, electrical, thermal, and mechanical properties, quantum dots have high fluorescence efficiency.86 Quantum dots can be used as signal reporters to achieve visual and fluorescent double signal amplification of probes, thereby enhancing the sensitivity of detection.
Quantum dots have numerous applications in exosome detection. For example, Boriachek’s group has proposed a voltammetric immunoassay that utilizes quantum dots as a signal amplifier for the electrochemical detection of tumor-specific exosomes. (Figure 5A).77 The assay consisted of three primary steps. In the first step, magnetic beads were employed to capture a large population of exosomes. This capture process was facilitated by utilizing cancer-specific antibodies, such as the CD63 antibody, which targets the tetraspanin protein commonly found on exosomes. Moving forward, specific cancer antibodies were utilized to specifically identify cancer-related exosomes. In this study, Biotinylated HER-2 and FAM134B antibodies were used to functionalize CdSe quantum dots, serving as specific markers for breast and colon cancer, respectively. After the identification step, magnetic washing and purification procedures were conducted out. Subsequently, CdSe quantum dots were dissolved using acid and then Cd2+ ions were quantified by anodic stripping voltammetry on a bare glassy carbon working electrode. The method is highly sensitive and can detect 100 exosomes/µL with a relative standard deviation (%RSD) of less than 5.5%. Furthermore, the dynamic range covered exosome concentrations from cancer cell lines (BT-474 and SW-48 cell lines) ranging from 102 to 107 particles/µL. This method uses quantum dots (QDs) to enhance the signal in stripping voltammetry immunoassay of tumor-specific exosomes. In the future, incorporating multiple quantum dots could lead to the creation of a single assay platform with the ability to simultaneously detect multiple tumor-specific exosomes. Ongoing research is focused on demonstrating exosomes detection in plasma, Vinduska’s group described a simple technique that employs quantum dots coupled with immunomagnetic capture and enrichment to detect exosome surface proteins. (Figure 5B).78 In this approach, the primary antibody specific to the target protein was used to recognize the surface protein markers on exosomes. Subsequently, a secondary antibody-conjugated quantum dot was employed to detect and label the exosomes. The detection process utilized fluorescent spectroscopy, taking advantage of the optical properties of the quantum dots. This approach based on quantum dots enabled highly sensitive detection of exosomes, with a LOD of 9.3×106 particles/mL. Notably, this LOD was more than 100 times lower than the typical concentration of exosomes found in plasma. Clinical studies revealed that exosomes derived from HER2-positive breast cancer patients exhibited approximately five times higher levels of HER2 expression compared to healthy controls, thus demonstrating their potential significance in clinical research and applications. Furthermore, quantum dots also showcased the capability to achieve sensitive detection of exosome miRNAs (exo-miRNAs). Li et al showcased a GQDs-functionalized reduced graphene oxide (RGO) field-effect transistor (FET) biosensor for the sensitive detection of exo-miRNAs in the blood of breast cancer patients (Figure 5C).79 To construct the biosensor, a combination of graphene quantum dots (GQDs) and amine-modified porous metal-organic frameworks (PMO) was formed by incubating them together. Subsequently, the RGO FET was fabricated, and polylysine (PLL) was assembled onto the surface of the RGO. This allowed for the immobilization of the GQDs-PMO hybrid on the chip surface through amide formation. Following immobilization, exosome-derived microRNAs (exo-miRNAs) were specifically detected by hybridizing the immobilized PMO probes with the miRNA. The interaction between exo-miRNAs and PMO on the chip surface led to a modification in the net carrier density of the sensing channel, attributable to the negative charges introduced by the exo-miRNAs. This shift in carrier density caused the Dirac point to shift towards the left. In summary, the sensing channel’s surface was modified with PMO probes, and the detection of miRNAs was achieved by monitoring the alteration in surface charge density during the PMO/miRNA hybridization process. This approach enabled the sensitive detection of exo-miR-21, with LOD of 2.04 fM. The distinctive three-dimensional structure of graphene quantum dots (GQDs) enhances hybridization efficiency, leading to improved detection performance of the GPPR-FET sensor. Moreover, this sensor can effectively differentiate variations in exo-miR-21 levels between breast cancer patients and healthy individuals, demonstrating its significant potential for clinical applications.
 |
Figure 5 (A) Schematic diagram of CdSe QDs based electrochemical strategy for exosomes detection. Reprinted from Boriachek K, Islam MN, Gopalan V, et al. Quantum dot-based sensitive detection of disease specific exosome in serum. Analyst. 2017;142(12):2211–2219. Copyright: ROyal SOciety of Chemistry, 2017.77 (B) Schematic mechanism of CdTe QDs based fluorescent system for exosomes assay. Reprinted from Vinduska V, Gallops CE, O’Connor R, et al. Exosomal surface protein detection with quantum dots and immunomagnetic capture for cancer detection. Nanomaterials. 2021;11(7):1853.78 (C) Schematic illustration of the GQDs-based electrochemical sensor for exosomes assay. Reprinted from Li K, Tu JY, Zhang YL, et al. Ultrasensitive detection of exosomal miRNA with PMO-graphene quantum dots-functionalized field-effect transistor biosensor. iScience. 2022;25(7):104522.79 |
Indeed, quantum dot materials have gained significant popularity in bio-sensing detection due to their various advantages:1) The fluorescence emission wavelength of quantum dots can be adjusted by altering their composition and particle size. Consequently, quantum dots with distinct fluorescence spectral characteristics can be synthesized. This unique property allows for the simultaneous study of multiple biological macromolecules in complex systems, which is challenging to achieve using traditional organic dye molecules under the same excitation wavelength. 2) Quantum dots exhibit narrow, symmetrical fluorescence spectral peaks. As biomarkers, they offer easily distinguishable fluorescence spectra with different characteristics, facilitating their identification and enhancing analysis. 3) Quantum dots demonstrate superior stability compared to organic dye molecules. They can withstand long-term and repeated light excitation without experiencing fluorescence bleaching. The combination of these factors in a single fluorescent probe is rare, positioning quantum dots as potential replacements for traditional molecular probes. Their adoption is expected to drive advancements in biological research and expand the application scope of quantum dot materials.
Au Nanomaterial Based Exosome Detection
Au nanoparticle with diameters ranging from 1–150 nm have unique optical, thermal, electrical, magnetism and chemical properties, which are strongly influenced by their size-dependent resonance absorption known as surface plasmon resonance (SPR) and Raman scattering (RS).87 These attributes render Au nanoparticles extremely beneficial in the realm of biosensors and biological sample analysis, particularly in the detection of exosomes.
To begin with, Gao’s group created a DNA-AuNP-based satellite network that combined low-speed centrifugal exosome isolation, detection, and protein analysis (Figure 6A).80 In this approach, a rolling circle amplification (RCA) reaction was utilized to generate a long-chain DNA hairpin structure. This hairpin structure comprised several functional domains, including CD63 aptamer sequences, linker sequences, and spacer sequences with complementary base pairs, resulting in the formation of a hairpin structure. Upon binding of exosomes to the CD63 aptamers on the DNA hairpin structure, the conformation of the hairpin structure was altered, exposing the linker sequences that contained binding sequences for gold nanoparticles (AuNPs). The probe molecules attached to the surface of the AuNPs then interacted with the long-chain DNA through a toehold-mediated strand displacement reaction. Consequently, the detection signal was the release of a fluorescent-labeled complementary probe, while also forming a satellite network based on DNA-AuNP. The method displayed a strong linear relationship between the change in fluorescence ratio (F/F0) and the logarithm of exosome concentration. The linear fit equation was F/F0=6.85lg Exo-33.51 with a high correlation coefficient (R2) of 0.9956. This approach utilizes a “two-mode” strategy for the detection and identification of exosomes. Initially, common fluorescence methods are employed to detect exosomes, followed by LC/MS techniques for protein profiling of the captured exosomes. This integration significantly enhances the accuracy of exosome detection and provides valuable insights for the development of “multi-mode” detection targets. In addition, in order to realize the detection of exosomes in complex samples, Wang’s group developed a method for detecting HIF-1α, an early biomarker for myocardial infarction, in circulating exosomes present in serum (Figure 6B).81 To achieve this, they employed gold nanospheres that were functionalized with a HIF-1α-binding aptamer by Au-S bonding. The gold seeds coated with aptamer-AuNP were grown using a seed-mediated growth method, which significantly enhanced the peroxidase-mimicking properties of the nanoparticles. This modification allowed for catalysis of a chromogenic reaction involving 3.3′5,5′-tetramethylbenzidine and hydrogen peroxide, resulting in improved sensitivity for detecting HIF-1α in circulating exosomes. The technique was particularly effective in rat serum samples induced with isoproterenol (ISO)-induced myocardial infarction. Notably, this strategy enabled the direct determination of HIF-1α in just a 25 μL sample, eliminating the need for preconcentration steps. This study effectively demonstrates the use of colorimetric nanoprobes to directly quantify exosome HIF-1α in serum samples from experimental myocardial infarction models. The pioneering strategy of using AuNPs-aptamer for ultra-low concentration analysis of exosome HIF-1α is employed to achieve high sensitivity detection in complex samples. Regarding the detection of exosome proteins, Cheng et al suggested a highly sensitive aptasensor that employs clover-like gold nanoclusters for the detection of breast exosomes. (Figure 6C).82 In this study, CD63 aptamer-modified DNA tetrahedrons were used as recognition elements for the specific exosome trapping on gold electrodes. Partially complementary DNA probes act as connectors to connect the captured exosomes to three AuNP-DNA signal probes. This novel clover-like structure was developed to overcome challenges in recognition and sensitivity caused by unwanted aggregation of AuNPs. When cancerous exosomes were present, a significant concentration of methylene blue molecules from DNA-AuNP nanocomposites on the electrode surface led to a robust current signal. In the concentration range of 1.0×103 to 1.0×108 particles/μL, there was a detectable response to MCF-7 cell-derived exosomes with a LOD of 158 particles/μL. Moreover, the aptasensor was successfully utilized for the analysis of serum samples obtained from breast cancer patients, showing excellent specificity. This sensor proved to be sensitive, simple, cost-effective, and suitable for widespread clinical application in tumor monitoring.
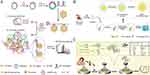 |
Figure 6 (A) Schematic illustration of the proposed DNA-AuNP-based satellite network structure for exosome analysis. Reprinted from Gao ML, Yin BC, Ye BC. Construction of a DNA-AuNP-based satellite network for exosome analysis. Analyst. 2019;144(20):5996–6003. Copyright: Royal Society of Chemistry, 2019.80 (B) Schematic illustration of aptamer-AuNP-coated gold seeds based colorimetric sensor for exosome detection. Reprinted from Wang QL, Huang WX, Zhang PJ, et al. Colorimetric determination of the early biomarker hypoxia-inducible factor-1 alpha (HIF-1α) in circulating exosomes by using a gold seed-coated with aptamer-functionalized Au@Au core-shell peroxidase mimic. Microchim Acta. 2019;187(1):61.81 (C) Schematic diagram of clover-like gold nanoclusters based aptasensor for exosomes detection. Reprinted from Cheng WT, Duan CJ, Chen Y, et al. Highly sensitive aptasensor for detecting cancerous exosomes based on clover-like gold nanoclusters. Anal Chem. 2023;95(7):3606–3612. Copyright: American Chemical Society, 2023.82 |
On the whole, the unique characteristic of Au nanomaterials is their ability to possess a wide range of core sizes ranging from 1 nm to 150 nm, making them easily modifiable with controlled dispersal. This renders GNPs highly promising for various biological detection applications. Unfortunately, the synthesis process of Au nanomaterials is susceptible to gold nanoparticle agglomeration, which can compromise the material’s performance. It is imperative to exercise strict control over the ratio of raw materials, temperature, time, and color changes during the synthesis process to ensure successful material synthesis. This, in turn, enhances both the stability of the material and biological analysis applicability.
GONs Material Based Exosome Detection
Graphene oxide (GO) is characterized by its brown and yellow color, while graphene, with its unique two-dimensional periodic honeycomb lattice structure, possesses distinct electronic and photoelectric properties.88 Graphene oxide nanomaterials exhibit light stability, good water solubility, excellent biocompatibility, and a surface that is easily chemically modified. Due to these characteristics, GO has promising applications in optics, materials science, and biological medicine analysis.
In the context of exosome detection, graphene oxide nanosheets play a vital role in target capture and amplification. Huang’s group has created magnetic graphene oxide nanoparticles (MGONs) for effective exosomes capturing (Figure 7A).83 Within this study, Fe3O4@SiO2 magnetic nanoparticles were functionalized with GO via dopamine, and the CD63 aptamers were subsequently attached to the surface of the resulting MGONs, enabling specific binding to CD63 proteins on the exosome membrane. The MGONs demonstrated superior exosome capture compared to Fe3O4@SiO2 alone, with 1.5 times more exosomes adsorbed onto their surface. Within 15 minutes at room temperature, the capture rate of exosomes is as high as 89.4%, and the exosomes could be efficiently enriched and isolated without impacting their morphological structure and biological functions under the action of external magnetic field. To quantify the captured exosomes, a FAM-labeled single-stranded DNA probes were hybridized with the MGONs, and fluorescence intensity was measured. This detection limit of exosomes, with a LOD of 2.4×107 particles/mL. Furthermore, these aptamer-decorated MGONs showed promise for exosomes capture in samples from HepG2 cancer patients. This sensor presents a promising solution for capturing exosomes with high efficiency. The aptamer-decorated MGONs used in this method have successfully captured exosomes from cancer patient samples, demonstrating the practical value of this approach for early clinical disease diagnosis. In order to further enhance the detection of exosomes with high sensitivity and selectivity, Lee et al have developed a strategy using silver nanoparticles (AgNP) and magnetic iron oxide NP (IONP) decorated on nanoparticle-decorated graphene (GRP) based biosensing platforms for detecting prostate-cancer-cell-derived exosome (PC-exosome) (Figure 7B).84 The hybrid nanomaterial consisting of AgNPs, IONP and GRP enabled the system to function as a magneto-plasmonic substrate for magnetofluoro-immunosensing (MFI) system. In this approach, the anti-prostate-specific antigen was immobilized on the Ag/IO-GRP and the PC-exosomes are separated from the sample using an external magnetic force. The introduction of dye-tetraspanin Ab induced the formation of a sandwich structure, and the resulting fluorescence intensity varied based on the number of exosomes present, leading to highly sensitive and selective detection performance. The method successfully detected PC-exosome in a culture media sample without prior purification, and the LOD is 134.32 particles/mL. The main advantage of this method is the development of the magnetofluoro-immunosensing (MFI) system. The MFI system demonstrates exceptional sensitivity and selectivity, making it highly promising for directly detecting exosomes in both culture media and clinical samples. This remarkable performance highlights the significant potential of the MFI system in advancing exosome detection methods for various research and clinical applications. Likewise, to broaden the linear range of exosome detection. In another study, Huang’s group have constructed a graphene-based biosensor capable of isolating breast cancer exosomes from extracellular vesicles (EVs) while enabling quantitative detection of exosomes (Figure 7C).85 The researchers utilized G-OH (hydroxylated graphene), a novel nanomaterial known for its exceptional properties such as high conductivity, good biocompatibility, and large surface area which make it ideal for biosensor preparation. The biosensor was constructed by depositing G-OH nanosheets to create a membrane with micropores that can selectively screen exosomes and microvesicles in extracellular vesicles (EVs). Subsequently, rabbit α-human CD44 antibody was incubated on the membrane to capture exosomes from MDA-MB-231 (MM-231). Lastly, the membrane was further modified with mouse α-human CD9 antibodies and a third anti-horseradish peroxidase complex to generate an electrochemical signal. The biosensor demonstrated a broad linear range of 25 to 1×106 particles/μL and a LOD of 9 particles/μL (S/N = 3), covering a span of 5 orders of magnitude. These findings indicate the potential for quantitatively detecting cancer cell exosomes. These investigations underscore the inventive application of graphene-based materials in biosensing platforms for identifying and isolating specific exosomes. As nanomaterial synthesis and characterization techniques continue to advance, the integration of plasmonic and magnetic properties, along with other unique characteristics of diverse nanomaterials, is anticipated to assume an increasingly crucial role in the sensitive and selective detection of various exosome types.
 |
Figure 7 (A) Schematic diagram of the aptamer-decorated MGONs structure for exosomes assay. Reprinted from Huang WG, Yu YR, Yang CY, et al. Aptamer decorated magnetic graphene oxide nanoparticles for effective capture of exosomes. Chem Eng J. 2022;431:133849.83 (B) Schematic diagram of Ag/IO-GRP-based MFI system for the exosomes detection. Reprinted from Lee J, Lee JH, Mondal J, et al. Magnetofluoro-immunosensing platform based on binary nanoparticle-decorated graphene for detection of cancer cell-derived exosomes. Int J Mol Sci. 2022;23(17):9619.84 (C) Schematic illustration of hydroxylated graphene based electrochemical sensors for exosomes detection. Reprinted from Huang J, Yan HD, Ma MX, et al. Hydroxylated graphene porous membrane-based biosensor for exosome isolation and detection. ACS Appl Nano Mater. 2022;5(5):6115–6124. Copyright: American CHemical Society, 2022.85 |
In summary, graphene oxide nanomaterials (GONs) are ideal substrate for capturing various biological molecules, such as exosomes, DNA, and cells, due to their exceptional physicochemical properties. This highlights their immense potential in the field of biomedical research. Combining GONs with emerging biosensor technologies, such as 3D printing, optical sensors, and electrochemical sensors, will yield significant advantages in biological detection as new technology advances and nanomaterials develop.
DNA Signal Amplification-Based Exosomes Detection
DNA signal amplification is a process that increases the detectable signal from DNA molecules to improve their detection sensitivity. Owing to its exceptional programmability, stability, and biocompatibility, the technique is highly valuable in a wide range of applications including molecular diagnostics, genetic testing, and biomedical research. Some notable examples include catalytic hairpin assembly (CHA), hybrid chain reaction (HCR), and rolling cycle amplification (RCA), etc.89 These techniques have proven especially useful in the field of exosome identification and detection. Table 4 presents a comprehensive overview of exosome detection methods that rely on DNA signal amplification.
 |
Table 4 An Overview on DNA Signal Amplification-Based Exosome Detection |
CHA Based Signal Amplification for Exosome Detection
CHA is an enzyme-free nucleic acid amplification technique that operates under isothermal conditions. It utilizes a unique free-energy-driven hairpin DNA assembly mechanism, offering several advantages such as low background noise, high turnover rates, and fast and effective signal amplification. CHA has found widespread use in exosome detection.101
For example, Zhang et al introduced a durable electrochemical biosensor for the analysis of exosomal miRNA, utilizing multifunctional DNA tetrahedrons assisted catalytic hairpin assembly (MDTs-CHA). (Figure 8A).90 The MDTs-CHA system incorporated two multifunctional tetrahedrons (T1 and T2) to facilitate rapid and highly sensitive analysis of exo-miRNAs through localized reactions and cascade amplification. Upon the presence of target exo-miRNA, the CHA reaction on DT1 was promptly initiated, leading to the exposure of numerous blocked P strands. This facilitated the specific capture of DT1 by the capture probe. Subsequently, the T1 strands on DT1 triggered the CHA reaction on DT2, resulting in the unfolding of blocked P strands on DT2, which were then captured by the electrode. Similarly, the T2 strands on DT2 could initiate the CHA reaction on T1. As a result, the electrode surface was coated with an ample number of DNA tetrahedrons, enabling the attachment of RuHex and resulting in the production of an electrochemical signal. This biosensor demonstrated the capacity to detect exosomal miRNA at concentrations as low as 7.2 aM within a 30 min. Furthermore, it demonstrated excellent performance with a high efficiency (AUC: 0.989) and a remarkable sensitivity of 90.5% in diagnosing breast tumors. This method uses DNA tetrahedron to amplify signals from the target, resulting in highly sensitive detection of exosome miRNA. This approach offers several advantages over traditional cascade amplification, including fewer reaction steps, straightforward operation, and improved specificity. These benefits make it a promising option for advancing exosome miRNA detection in research and clinical settings. In order to enhance the versatility of assay methods for exosomes originating from diverse cell types. Zhou’s group introduced a technique known as aptamer-initiated catalytic hairpin assembly (AICHA) fluorescence assay for the highly sensitive identification of cancer-derived exosomes. (Figure 8B).91 The assay employed aptamers that were specifically designed to target proteins and biotin modified, with a partial blocking by initiators. These modified aptamers were then conjugated onto streptavidin-modified magnetic beads (SA-MB) through the interaction between biotin and streptavidin. The aptamers could selectively recognize and capture exosomes by binding to the membrane proteins on their surface, causing the initiators to be released into the supernatant. Following magnetic separation, the free initiators initiated the CHA circuit. Subsequently, the released initiator sequentially triggered the next CHA circuit, leading to the formation of numerous H1-H2 duplexes and resulting in amplification of the FAM fluorescence signal. The efficacy of the AICHA method was confirmed by successfully detecting exosomes derived from MCF-7 cells, showcasing a wide calibration range of 8.4 particles/μL to 8.4×105 particles/μL and a low LOD of 0.5 particles/μL. Furthermore, the versatility of the AICHA approach was demonstrated by detecting exosomes derived from PANC-1 cells, with an LOD of 0.1 particles/μL. The AICHA platform robustly applies to the analysis of exosome protein biomarkers and aids in clinical diagnosis. Its adaptability to various applications and ability to deliver reliable results enhances our understanding of exosomes and their potential role in diagnostics and therapeutics. Regarding the detection of piRNAs, in a study by Zhang’s group, a novel method called universal catalytic hybridization assembly system (uniCHA) was developed to for the quantification of exosomal piRNAs as well as microRNAs (Figure 8C).92 The uniCHA system was comprised of three hairpin probes: H1 labeled with Cy3, H2 labeled with Cy5, and an initial hairpin H0 with a partially fixed sequence. H0 consisted of three functional parts: 1, 2, and 3. Sequence 1 acted as the trigger to open hairpin H1, sequence 2 was complementary to the target miRNA or piRNA, and sequence 3 formed the stem of H0. To conduct the assay, the three hairpin probes were introduced into a pre-treated plasma sample containing minute quantities of piRNA or miRNA from ruptured exosomes. The target molecules hybridized with the corresponding loop part in H0, leading to the unfolding of the hairpin structures. This initiated the subsequent H1-H2 assembly amplification circuit. The concentration of the assembled product H1-H2 at a specific timepoint before reaching equilibrium was directly proportional to the original target quantity. Quantification was accomplished by measuring the fluorescence resonance energy transfer (FRET) signal resulting from the proximity of Cy3 and Cy5 in the product. The uniCHA system facilitated the detection and analysis of diverse piRNAs and miRNAs under consistent reaction conditions with minimal background noise and high sensitivity at the picomolar level. This system successfully detected piR-651 and miR-1246 in exosomes secreted by MCF-7 cells at a concentration of 106 particles/μL. Additionally, they conducted a direct plasma biopsy to diagnose breast cancer and achieved a 100% sensitivity and specificity in a group of 21 breast cancer patients and 13 healthy controls. The uniCHA system is an exceptional method for quantifying piRNAs. This is a significant breakthrough as current methods for analyzing piRNAs are limited to reverse transcription quantitative polymerase chain reaction (RT-qPCR) and next-generation sequencing (NGS). The uniCHA system is a valuable addition to the existing techniques, offering researchers an alternative approach for precise quantification of piRNAs. Similarly, for miRNA detection, in another study established by Wu’s group, a biosensor utilizing multi-branched localized catalytic hairpin assembly (MLCHA) and photonic crystals (PCs) was developed for the on-site detection of exosomal miRNA. (Figure 8D).93 The MLCHA probes were composed of five single-stranded DNAs (S1-S5) and a pair of hairpin probes (H1 and H2) on each branch. These probes exhibited excellent stability and could enter exosomes without causing damage. When the target miRNA-27a was recognized, the self-quenched hairpin probe of H1 unfolded, resulting in fluorescence recovery. The freshly exposed DNA segment of H1 subsequently initiated the unwinding of H2, leading to the formation of a thermodynamically favorable H1/H2 duplex and the release of the miRNA for a new cycle. The process generated an amplified fluorescence signal because a single target miRNA could open multiple hairpin probes of H1. The fluorescence signal was further enhanced by the presence of PCs. The biochip strategy proposed was utilized to detect exosomal miRNA-27a across a range of concentrations, demonstrating a linear relationship from 0.76×109 particles/mL to 15.24×109 particles/mL with a correlation coefficient (R2) of 0.99. The LOD was calculated to be 1.9×107 particles/mL. The biosensor, developed using MLCHA and PCs, eliminates the need for time-consuming and laborious steps such as exosome lysis and RNA extraction, guaranteeing assay accuracy. Additionally, the remarkable rigidity and stability of MLCHA probes make this approach applicable to non-invasive liquid biopsy and the detection of tumor biomarkers.
In summary, CHA demonstrates high sensitivity and can aid in the detection of exosomal protein and miRNA target sequences. However, due to the spontaneous breathing process of DNA duplexes, CHA hairpins may generate non-specific reactions in the absence of an initiator, resulting in elevated background signals. To reduce these signals, efforts should be directed towards rational sequence design to prevent unexpected reactions and improve detection accuracy.
 |
Figure 8 (A) Schematic diagram of MDTs-CHA based electrochemical platform for exo-miRNA detection. Reprinted from Zhang Y, Zhang XH, Situ B, et al. Rapid electrochemical biosensor for sensitive profiling of exosomal microRNA based on multifunctional DNA tetrahedron assisted catalytic hairpin assembly. Biosens Bioelectron. 2021;183:113205.90 (B) Principle of the proposed AICHA based signal amplification strategy for exosome detection. Reprinted from Zhou JQ, Lin QY, Huang ZP, et al. Aptamer-initiated catalytic hairpin assembly fluorescence assay for universal, sensitive exosome detection. Anal Chem. 2022;94:5723–5728. Copyright: AMerican Chemical Society, 2022.91 (C) Schematic illustration of the universal CHA system based signal amplification strategy for detecting exosomes. Reprinted from Zhang LM, Gao QX, Chen J, et al. A universal catalytic hairpin assembly system for direct plasma biopsy of exosomal PIWI-interacting RNAs and microRNAs. Anal Chim Acta. 2022;1192:339382.92 (D) Schematic illustration of MLCHA and PCs based sensing biochip for the in situ exo-miRNA detection. Reprinted from Wu TT, Liu XS, Chen HJ, et al. An in situ exosomal miRNA sensing biochip based on multi-branched localized catalytic hairpin assembly and photonic crystals. Biosens Bioelectron. 2023;222:115013.93 |
HCR Based Signal Amplification for Exosome Detection
Hybrid chain reaction (HCR) is a DNA amplification process that involves the addition of specific DNA strands that act as triggers to initiate a chain reaction. This process leads to the opening and subsequent hybridization of two hairpin DNA structures, forming a long double-stranded DNA (dsDNA) molecule. The HCR process can be performed under mild conditions and does not require the presence of any specific species or organisms. This makes it a versatile and robust amplification technology. It offers advantages such as signal amplification, which enables sensitive target detection. Due to its ease of use and constant temperature requirements, HCR has gained significant attention in practical analysis applications. Its simplicity of operation makes it an attractive choice for researchers and practitioners in various fields, including molecular biology and diagnostics.
A study by Zhang et al demonstrated the application of HCR in highly sensitive detection of exosome. In this approach, HCR was used to amplify the signal by introducing more alkaline phosphatase (ALP) enzymes (Figure 9A).94 To begin the process, exosomes are captured using CD63 aptamers labeled with magnetic beads. DNA probes modified with cholesterol are then inserted into the lipid membrane of the exosomes. The ends of these DNA probes act as initiators to trigger a hybridization chain reaction (HCR), leading to signal amplification. The HCR process increases the number of sites available for loading alkaline phosphatase (ALP), which enhances the generation of ascorbic acid. Ascorbic acid then reduces silver ions, resulting in the formation of silver shells on gold nanorods (Au NRs), causing a blue shift in the longitudinal localized surface plasmon resonance peak. The concentration of exosomes can be visually distinguished by the naked eye due to the vivid color variation caused by the formation of the silver shells. This method demonstrates detection limits as low as 1.6×102 particles/μL using UV-vis spectroscopy and 9×103 particles/μL by visual observation. Compared to other colorimetric methods for exosome quantification, this approach offers the advantage of visualizing results through a variety of distinct color tones. Utilizing Au NRs as signal reporters, this method introduces a unique and visually striking color change that enables the visual analysis of exosomes. This approach provides a simple, immediate, and efficient way to detect exosomes without requiring complex and expensive equipment, offering researchers a highly accessible tool for rapid and reliable analysis of these important biomolecules. In terms of the detection of exosomes in clinical samples, Zhang’s group introduced an electrochemical micro-aptasensor for the highly sensitive detection of exosomes. This approach involved the integration of a micropatterned electrochemical aptasensor with a hybridization chain reaction (HCR) signal amplification method. (Figure 9B).95 In this method, exosomes are specifically enriched on electrodes functionalized with CD63 aptamers. The enriched exosomes are then recognized by hybridization chain reaction (HCR) products, which consist of avidin-horseradish peroxidase (HRP) complexed with EpCAM aptamers as bridges. The current signal generated by the enzyme reaction between HRP and 3.3’, 5.5’-tetramethylbenzidine (TMB)/H2O2 directly corresponds to the amount of bound HRP on the HCR products, providing a measurement of the target exosome quantity. To enhance specificity, anti-EpCAM aptamers are introduced into the micro-aptasensors, allowing for the selective detection of cancerous exosomes. This dual-amplification strategy enables a linear detection response over a broad range of exosome concentrations, from 2.5×103 to 1×107 particles/mL. The detection limit achieved was 5×102 particles/mL. The HCR-based microelectrodes signal sensors have effectively detected cancerous exosomes in serum samples from both early-stage and late-stage lung cancer patients. This breakthrough offers valuable insights into early cancer diagnosis, providing a promising avenue for the early detection and treatment of lung cancer. In terms of the detection of exosomal miRNA in clinical samples, Kim et al proposed a hydrogel-based HCR for multiplex signal amplification to detect urinary exosomal miRNAs from human clinical samples (Figure 9C).96 This method comprises three primary stages: target hybridization, initiation, and HCR amplification. The hydrogel-based HCR process commences with the binding of universal initiators, which primes the probe regions of the hydrogel to initiate the HCR process. The central process of HCR is initiated when a universal pair of biotinylated hairpins (hairpins 1 and 2) hybridize with the probe regions of the hydrogel. Subsequently, Hairpin 1, which is complementary to the universal initiator, sequentially unwinds its remaining sequences, exposing biotin. A similar hybridization process occurs with Hairpin 2, leaving sequences complementary to hairpin 1 and free of biotin. This chain reaction continues, leading to the accumulation of multiple biotins within the hydrogel until the HCR is terminated. Finally, phycoerythrin-conjugated streptavidin (SA-PE) is introduced to the biotins of the hairpins for fluorescent reporting, enhancing the fluorescence readout signal for each target miRNA’s hybridization event. The hydrogel-based HCR method can identify urinary exosomal miRNAs from clinical human samples as a non-invasive diagnostic tool for prostate cancer. According to the researchers, this study was the first to analyze exosomal miRNAs without target amplification using less than 1 mL of urine. The technique allows the amplification, approximately 35-fold and successfully detected two urinary exosomal miRNAs (hsa-miR-6090 and hsa-miR-3665) from 19 patients with prostate cancer and 19 healthy controls from clinical samples (600 μL/sample). The platform has been validated for its potential clinical applicability, eliminating the need for normalization to an endogenous reference gene. This valuable tool can complement the existing PSA test for prostate cancer screenings. By offering an alternative approach, this platform opens up new possibilities for more accurate and comprehensive diagnosis of prostate cancer. In relation to the research of specific glycosylation of exosomal proteins, Kang’s group used induced HCR to develop a proximity dual-tagging strategy for amplified visualization and functional exploration of exosomal protein-specific glycosylation (Figure 9D).97 Exosomal metabolic glycan labeling is achieved through cellular MGL, and glycans are efficiently labeled as an associative activation toehold (DBCO-T1) for glycan signal transduction through subsequent copper-free bioorthogonal reactions. The target protein on the exosome is recognized by an aptamer probe (Aptamer-T2), which consists of an aptamer targeting the non-functional epitope of the target protein and another activation toehold. When DBCO-T1 and Aptamer-T2 combined into the same exosomal glycoprotein, their proximity induces the formation of an associative toehold nanostructure. The hairpin structure of fluorescently labeled H1 enables the HCR assembly of H1 and H2 probes, resulting in the amplification of the signal for exosomal protein-specific glycosylation. It is worth noting that compared with no HCR amplification of signal, the method will secrete body outside the PD-L1 (exoPD-L1) visualization of increased 7.7 times, highlights the secrete body outside the study the efficacy of glycoprotein. Importantly, this amplification does not interfere with the natural exoPD-L1/PD-1 interaction, making it a powerful tool for studying the biological implications of exosomal glycoproteins.
HCR, a nucleic acid amplification technology, has been extensively used for exosome detection due to its highly sensitive and precise outcomes. The localized HCR approach offers a faster reaction time by utilizing the spatial-confinement effect and therefore represents a promising alternative to the limitations of random hairpin collisions.
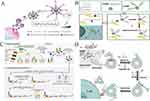 |
Figure 9 (A) Schematic illustration of the HCR based signal amplification strategy for exosome detection. Reprinted from Zhang YZ, Wang D, Yue S, et al. Sensitive multicolor visual detection of exosomes via dual signal amplification strategy of enzyme-catalyzed metallization of Au nanorods and hybridization chain reaction. ACS Sens. 2019;4(12):3210–3218. Copyright: American Chemical Society, 2019.94 (B) Schematic mechanism of the HCR based electrochemical micro-aptasensors for exosome detection. Reprinted from Zhang WF, Tian ZH, Yang SJ, et al. Electrochemical micro-aptasensors for exosome detection based on hybridization chain reaction amplification. Microsyst Nanoeng. 2021;7(1):63.95 (C) Schematic diagram of hydrogel-based HCR signal amplification procedure for exosome assay. Reprinted from Kim J, Shim JS, Han BH, et al. Hydrogel-based hybridization chain reaction (HCR) for detection of urinary exosomal miRNAs as a diagnostic tool of prostate cancer. Biosens Bioelectron. 2021;192:113504. Creative Commons.96 (D) Schematic diagram of HCR strategy for characterizing exoPD-L1 glycosylation. Reprinted from Kang SY, Zhu L, Wang WC, et al. Amplified visualization and function exploration of exosomal protein-specific glycosylation using hybridization chain reaction from non-functional epitope. Sci China Chem. 2022;65(6):1204–1211.97 |
RCA Based Signal Amplification for Exosome Detection
Rolling circle amplification (RCA) is a technique involving the continuous amplification of DNA under temperature control, utilizing a DNA polymerase. In this technique, oligonucleotides labeled with antibodies were efficiently amplified up to 109 times. The amplified product underwent complementary hybridization with subsequently added oligonucleotides labeled with tracers, resulting in the generation of numerous hybrids between the labeled tracers and the antigen-antibody complexes.102 Ultimately, the tracer was amplified, further bolstering the signal. This approach offers high sensitivity and specificity, allowing for the detection of extremely low antigen concentrations by amplifying the signal through multiple rounds of hybridization and amplification. Such an approach can be highly valuable in various applications where the sensitive detection of antigens is crucial.
RCA is a crucial component in the detection of exosomes. For instance, Sun et al developed a universal target-initiated proximity ligation assay (TIPLA) for the sensitive detection of miRNA by combining target miRNA initiation and proximity ligation-triggered RCA. (Figure 10A).98 In the TIPLA technique, miRNA unfolds H1 probes that are tethered to the surface of streptavidin magnetic beads (SMBs) to initiate the entire assay. Subsequently, the miRNA is displaced for the next catalytic cycle (primary amplification) through the interaction between H1 and a Connector. Following this, the proximity ligation induces the generation of single-strand DNA (ssDNA), which is detected by a fluorescence probe and enriched using magnets. This method demonstrates excellent sensitivity with a LOD of 30.25 fM. This method directly detects trace exosomal miRNAs without the need for an additional reverse transcription procedure. It offers a novel avenue for bioanalysis in living systems and holds great potential for contributing to the diagnosis of cardiovascular disease. Wang’s group has also reported a sensitive method for detecting multiple exosomal miRNAs using RCA (Figure 10B).99 In this assay, three specific miRNAs, namely miRNA-21 (mir21), miRNA-155 (mir155), and miRNA-122 (mir122), were selected as model miRNAs. Three corresponding padlock probes targeting these miRNAs were introduced, along with T4 DNA ligase, to form circular DNA templates. Subsequent simultaneous RCA was initiated by adding phi29 DNA polymerase and dNTPs. Molecule beacons (MBs) were then introduced to detect the RCA products. Each MB consisted of a loop and two complementary stems, with a fluorophore and a quencher modified at each end. Before hybridization, the Molecular Beacons (MBs) adopted a hairpin structure with quenched fluorophores. Upon binding with the target miRNA, the MBs hybridized with it, leading to the opening of the hairpin structure and fluorescence recovery. The fluorescence intensity ratio of the three MBs provided valuable information about the abundance of the three exosomal miRNAs, enabling profiling and screening of exosomal miRNAs. The LOD for miRNA-122, miRNA-155, and miRNA-21 were determined to be 1.16 fM, 1.36 fM, and 8.84 fM, respectively. Compared to the traditional PCR method, multiplexed RCA detection provides similar results in less time and with fewer resources. Additionally, the RCA process improves the resolution of the method compared to simple fluorescence detection. The multiplexed RCA detection method represents a significant improvement in molecular diagnostics and holds great promise for various applications in research and clinical settings. Regarding the detection of exosome protein, a dual-signal amplified colorimetric biosensor for the detection of exosomes was constructed by Li’s group, through the integration of rolling circle amplification (RCA) and gold nanoparticles (GNPs). (Figure 10C).100 In this study, nucleolin, a biomarker identified on exosomes derived from leukemia cells, was targeted for detection. The immobilization of the complex formed by a trigger probe and an aptamer probe was achieved using magnetic beads (MBs). Upon the introduction of exosomes, the high affinity between the aptamer and nucleolin resulted in the release of the trigger probe. This triggered an RCA reaction on the magnetic beads. This process generated multiple repeated sequences that hybridized with the signal probe. Subsequently, GNPs-HRP bound to the signal probe triggered a color change in the presence of tetramethylbenzidine (TMB). As a result, this visual color change enabled highly sensitive detection of exosomes, with a LOD of 100 particles/μL. The method demonstrated strong linearity (R2 = 0.9944), effectively distinguishing healthy individuals from leukemia patients. This underscores its potential clinical utility in liquid biopsy for leukemia. The accurate differentiation between healthy controls and leukemia patients using this method provides hope for improved diagnosis and monitoring of the disease. This method demonstrates the potential to advance liquid biopsy as a valuable tool in leukemia diagnosis and management, given its robust performance and significant clinical benefits.
The RCA reaction provides a remarkable amplification of over a billion-fold within 1–2 h at room temperature, positioning it as a widely used method for the detection of exosomes and complex biological samples. Despite the persistent challenges of specificity and sensitivity, the RCA reaction remains a promising technology for detection. To maximize hybridization efficiency and enhance detection sensitivity while eliminating non-specific templates in the detection environment, it is recommended to conduct pre-tests with optimized conditions to determine the hybridization dynamic curve between different nucleic acid chains prior to experimentation.
 |
Figure 10 (A) Working principle of the proposed RCA strategy for exo-miRNA detection. Reprinted from Sun X, Chen J, Lang JL. Sensitive detection of exosomal miRNA for cardiovascular diseases with target initiate proximity ligation assay (TIPLA). Microchem J. 2020;158:105193.98 (B) Schematic illustration of RCA signal amplification strategy for multiple exo-miRNAs. Reprinted from Wang ZL, Zong SF, Liu Y, et al. Simultaneous detection of multiple exosomal microRNAs for exosome screening based on rolling circle amplification. Nanotechnology. 2021;32(8):085504.99 (C) Schematic diagram of RCA induced dual signal amplification platform for exosomes detection. Reprinted from Li C, Zhou MY, Wang HY, et al. Rolling circle amplification assisted dual signal amplification colorimetric biosensor for ultrasensitive detection of leukemia-derived exosomes. Talanta. 2022;245:123444.100 |
Conclusions and Outlook
Exosomes play a crucial role in facilitating communication between tumors and their target cells. It is of utmost importance to accurately isolate and detect exosomes for various purposes, including early diagnosis and monitoring prognosis. In this review, we provide a comprehensive summary of different isolation techniques, as well as nanomaterials-based and DNA signal amplification techniques, for the detection of exosome. Nanomaterials possess unique characteristics that make them highly attractive for signal transduction and the development of biosensors. Moreover, the incorporation of DNA signal amplification strategies has notably improved the sensitivity of biosensors for exosome detection. Despite the substantial advancements in exosome detection, there remain several challenges that require attention. 1) accurately identifying disease-specific exosomes in complex backgrounds of real samples remains a primary technical challenge, particularly when dealing with interference from other biomarkers present inside tumor cells. 2) the presence of various nucleases in biological samples, including exosomes, can decrease the stability of nanomaterial probes, making it challenging to achieve continuous detection of target molecules. 3) the issue of high background noise caused by non-specific hybridization between DNA molecules still persists in DNA-based signal amplification technologies.
Despite the existing challenges in exosome detection, efforts can be made to solve these problems from the following points.1) Accurate design of aptamer sequences: In detection methods based on Aptamer recognition, it is crucial to precisely design the sequence of aptamers to ensure their effectiveness in targeting specific exosomal proteins. 2) Modification of nanomaterials: Covalent or noncovalent modifications on nanomaterials can be explored to improve their stability while maintaining their detection capabilities. It is important to minimize any impact on the performance of nanomaterials during modification. 3) Rational design of DNA probes: In DNA signal amplification technology, precise synthesis of DNA sequences and flexible construction of DNA microstructures can improve the practicality of DNA probes. Additionally, alternative means of signal readout such as electrochemical biosensors can be introduced to enhance detection efficiency. 4) Simultaneous detection of multiple biomarkers: Relying solely on a single biomarker may not accurately reflect the stage of disease due to individual differences in disease progression. Therefore, the rational design of probes can enable the simultaneous detection of several disease-associated biomarkers in exosomes, providing a more comprehensive understanding of the disease.
We are confident that the continuous efforts to enhance exosome detection techniques will lead to the creation of an optimal next-generation platform. This progress empowers us to employ more advanced design and synthetic techniques for tailoring exosome-based theranostic nanoplatforms. The ongoing advancements in nanoscience and nanotechnology within material science, chemistry, biology, oncology, and pharmacy will undoubtedly have a significant impact on optimizing early cancer detection, improving therapeutic efficacy. This platform will be suitable for routine exosome analysis in both research and clinical settings, and will play a significant role in promoting persistent innovations within the scientific disciplines of nanomedicine. Furthermore, it will break through the limitations currently impeding the further advancement of theranostic nanomedicine, making a valuable contribution to the field.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 82074122 and 82304890) and Research Foundation of Education Bureau of Hunan Province (Grant No. 23A0280) and Key Research and Development Programs of Hunan Science and Technology Department (Grant No. 2023SK2046) and Opening Funding of Traditional Chinese Medicine and Ethnomedicine Innovation & Development International Laboratory (Grant No. 2022GJSYS04 and 2022GJSYS07) and Changsha Natural Science Foundation (Grant No. Kq2208193).
Disclosure
Qiongdan Zhang and Huizhen Wang are co-first authors for this study. The authors declare no conflicts of interest in this work.
References
1. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–383. doi:10.1083/jcb.201211138
2. Siravegna G, Marsoni S, Siena S, et al. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017;14(9):531–548. doi:10.1038/nrclinonc.2017.14
3. Wang J, Cheng S, Li G, et al. Application of liquid biopsy in precision medicine: opportunities and challenges. Front Med. 2017;11(4):522–527. doi:10.1007/s11684-017-0526-7
4. Chit C, Ka FT. Latest development of liquid biopsy. J Thorac Dis. 2018;10(S14):S1645–S1651. doi:10.21037/jtd.2018.04.68
5. Kallur R. The biology and function of exosomes in cancer. J Clin Invest. 2016;126(4):1208–1215. doi:10.1172/JCI81135
6. Lim CZJ, Zhang L, Zhang Y, et al. New sensors for extracellular vesicles insights on constituent and associated biomarkers. ACS Sens. 2020;5(1):4–12. doi:10.1021/acssensors.9b02165
7. Yang B, Chen Y, Shi JL. Exosome biochemistry and advanced nanotechnology for next-generation theranostic platforms. Adv Mater. 2019;31:2.
8. Shao HL, Im H, Castro CM, et al. New technologies for analysis of extracellular vesicles. Chem Rev. 2018;118(4):1917–1950. doi:10.1021/acs.chemrev.7b00534
9. Wei H, Chen Q, Lin L, et al. Regulation of exosome production and cargo sorting. Int J Biol Sci. 2021;17(1):163–177. doi:10.7150/ijbs.53671
10. Meng XD, Müller V, Milde LK, et al. Diagnostic and prognostic relevance of circulating exosomal miR-373, miR-200a, miR-200b and miR-200c in patients with epithelial ovarian cancer. Oncotarget. 2016;7(13):16923–16935. doi:10.18632/oncotarget.7850
11. Chang KL, Sun P, Dong X, et al. Aptamers as recognition elements for electrochemical detection of exosomes. Chem Res Chin Univ. 2022;38(4):879–885. doi:10.1007/s40242-022-2088-8
12. Wiseman JM, Evans CA, Bowen CL, et al. Direct analysis of dried blood spots utilizing desorption electrospray ionization (DESI) mass spectrometry. Analyst. 2010;135(4):720–725. doi:10.1039/b922329k
13. Lötvall J, Hill AF, Hochberg F, et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the international society for extracellular vesicles. J Extracell Vesicles. 2014;3(1):26913. doi:10.3402/jev.v3.26913
14. Swathi M, Tushar P. Extracellular vesicle long noncoding RNA as potential biomarkers of liver cancer. Brief Func Genomics. 2016;15(3):249–256. doi:10.1093/bfgp/elv058
15. Verweij FJ, Van Eijndhoven MAJ, Hopmans ES, et al. LMP1 association with CD63 in endosomes and secretion via exosomes limits constitutive NF-κB activation. EMBO J. 2011;30(11):2115–2129. doi:10.1038/emboj.2011.123
16. György B, Szabó TG, Pásztói M, et al. Membrane vesicles, current state-of-the-art emerging role of extracellular vesicles. Cell Mol Life Sci. 2011;68(16):2667–2688. doi:10.1007/s00018-011-0689-3
17. Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73(10):1907–1920. doi:10.1016/j.jprot.2010.06.006
18. Dignat-George F, Boulanger CM. The many faces of endothelial microparticles. Arterioscler Thromb Vasc Biol. 2011;31(1):27–33. doi:10.1161/ATVBAHA.110.218123
19. Zeng ZC, Li YL, Pan YJ, et al. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat Commun. 2018;9(1):5395. doi:10.1038/s41467-018-07810-w
20. He Q, Ye A, Ye W, et al. Cancer-secreted exosomal miR-21−5p induces angiogenesis and vascular permeability by targeting KRIT1. Cell Death Dis. 2021;12(6):576. doi:10.1038/s41419-021-03803-8
21. Liang GF, Zhu YL, Ali DJ, et al. Engineered exosomes for targeted co-delivery of miR-21 inhibitor and chemotherapeutics to reverse drug resistance in colon cancer. J Nanobiotechnology. 2020;18(1):10. doi:10.1186/s12951-019-0563-2
22. Ma ZJ, Wei K, Yang FM, et al. Tumor-derived exosomal miR-3157-3p promotes angiogenesis, vascular permeability and metastasis by targeting TIMP/KLF2 in non-small cell lung cancer. Cell Death Dis. 2021;12(9):840. doi:10.1038/s41419-021-04037-4
23. Sandfeld PB, Aggerholm PN, Bæk R, et al. Exosomal proteins as prognostic biomarkers in non-small cell lung cancer. Mol Oncol. 2016;10(10):1595–1602. doi:10.1016/j.molonc.2016.10.003
24. Huang SH, Li Y, Zhang J, et al. Epidermal growth factor receptor-containing exosomes induce tumor-specific regulatory T cells. Cancer Invest. 2013;31(5):330–335. doi:10.3109/07357907.2013.789905
25. Yao XL, Tu Y, Xu YL, et al. Endoplasmic reticulum stress-induced exosomal miR-27a-3p promotes immune escape in breast cancer via regulating PD-L1 expression in macrophages. J Cell Mol Med. 2020;24(17):9560–9573. doi:10.1111/jcmm.15367
26. Soo CY, Song YQ, Zhen Y, et al. Nanoparticle tracking analysis monitors microvesicle and exosome secretion from immune cells. Immunology. 2012;136(2):192–197. doi:10.1111/j.1365-2567.2012.03569.x
27. Filipe V, Hawe A, Jiskoot W. Critical evaluation of nanoparticle tracking analysis (NTA) by nanosight for the measurement of nanoparticles and protein aggregates. Pharm Res. 2010;27(5):796–810. doi:10.1007/s11095-010-0073-2
28. Liu CC, Xu XN, Li B, et al. Single-Exosome-Counting Immunoassays for Cancer Diagnostics. Nano Lett. 2018;18(7):4226–4232. doi:10.1021/acs.nanolett.8b01184
29. Lee J, Kim H, Heo Y, et al. Enhanced paper-based ELISA for simultaneous EVs/exosome isolation and detection using streptavidin agarose-based immobilization. Analyst. 2019;145(1):157–164. doi:10.1039/C9AN01140D
30. Hsu CC, Wu Y. Recent advances in nanotechnology-enabled biosensors for detection of exosomes as new cancer liquid biopsy. Exp Biol Med. 2022;247(23):2152–2172. doi:10.1177/15353702221110813
31. Gurunathan S, Kang MH, Kim JH. A comprehensive review on factors influences biogenesis, functions, therapeutic and clinical implications of exosomes. Int J Nanomed. 2021;16:1281–1312. doi:10.2147/IJN.S291956
32. Saad MG, Beyenal H, Dong WJ. Exosomes as powerful engines in cancer isolation, characterization and detection techniques. Biosensors. 2021;11(12):518. doi:10.3390/bios11120518
33. Ma XH, Hao YQ, Liu L. Progress in nanomaterials-based optical and electrochemical methods for the assays of exosomes. Int J Nanomed. 2021;16:7575–7608. doi:10.2147/IJN.S333969
34. Zhao Z, Fan J, Hsu YM, Lyon CJ, Ning B, Hu TY. Extracellular vesicles as cancer liquid biopsies from discovery, validation, to clinical application. Lab Chip. 2019;19(7):1114–1140. doi:10.1039/C8LC01123K
35. Pang B, Zhu Y, Ni J, et al. Extracellular vesicles the next generation of biomarkers for liquid biopsy-based prostate cancer diagnosis. Theranostics. 2020;10(5):2309–2326. doi:10.7150/thno.39486
36. Garcia-Romero N, Esteban-Rubio S, Rackov G, et al. Extracellular vesicles compartment in liquid biopsies clinical application. Mol Aspects Med. 2018;60:27–37. doi:10.1016/j.mam.2017.11.009
37. Liu JL, Chen Y, Pei F, et al. Extracellular vesicles in liquid biopsies potential for disease diagnosis. Biomed Res Int. 2021;2021:6611244. doi:10.1155/2021/6611244
38. Xu K, Jin YL, Li YM, et al. Recent progress of exosome isolation and peptide recognition-guided strategies for exosome research. Front Chem. 2022;10:844124. doi:10.3389/fchem.2022.844124
39. Lai JJ, Chau ZL, Chen SY, et al. Exosome processing and characterization approaches for research and technology development. Adv Sci. 2022;9(15):e2103222. doi:10.1002/advs.202103222
40. Li P, Kaslan M, Lee SH, et al. Progress in exosome isolation techniques. Theranostics. 2017;7(3):789–804. doi:10.7150/thno.18133
41. Jiang ZJ, Liu GY, Li JP. Recent progress on the isolation and detection methods of exosomes. Chem Asian J. 2020;15(23):3973–3982. doi:10.1002/asia.202000873
42. Hu PY, Wang X, Wei L, et al. Selective recognition of CdTe QDs and strand displacement signal amplification-assisted label-free and homogeneous fluorescence assay of nucleic acid and protein. J Mater Chem B. 2019;7(31):4778–4783. doi:10.1039/C9TB00753A
43. Chia BS, Low YP, Wang Q, et al. Advances in exosome quantification techniques. Trends Anal Chem. 2017;86:93–106. doi:10.1016/j.trac.2016.10.012
44. Lin SJ, Yu ZX, Chen D, et al. Progress in microfluidics-based exosome separation and detection technologies for diagnostic applications. Small. 2020;16(9):e1903916. doi:10.1002/smll.201903916
45. Mahgoub EO, Razmara E, Bitaraf A, et al. Advances of exosome isolation techniques in lung cancer. Mol Biol Rep. 2020;47(9):7229–7251. doi:10.1007/s11033-020-05715-w
46. Zhang Y, Bi JY, Huang JY, et al. Exosome: a review of its classification, isolation techniques, storage, diagnostic and targeted therapy applications. Int J Nanomed. 2020;15:6917–6934.
47. Boriachek K, Islam MN, Möller A, et al. Biological functions and current advances in isolation and detection strategies for exosome nanovesicles. Small. 2018;14(6):6. doi:10.1002/smll.201702153
48. Xu WM, Li A, Chen JJ, et al. Research development on exosome separation technology. J Membr Biol. 2023;256(1):25–34. doi:10.1007/s00232-022-00260-y
49. Ludwig N, Whiteside TL, Reichert TE. Challenges in exosome isolation and analysis in health and disease. Int J Mol Sci. 2019;20(19):4684. doi:10.3390/ijms20194684
50. Le N, Fan ZH. Exosome isolation using nanostructures and microfluidic devices. Biomed Mater. 2021;16(2):022005. doi:10.1088/1748-605X/abde70
51. Wang J, Ma P, Kim DH, et al. Towards microfluidic-based exosome isolation and detection for tumor therapy. Nano Today. 2021;37:101066. doi:10.1016/j.nantod.2020.101066
52. Bu HC, He DG, He XX, et al. Exosomes: isolation, analysis, and applications in cancer detection and therapy. Chembiochem. 2019;20(4):451–461. doi:10.1002/cbic.201800470
53. Cho S, Yang HC, Rhee WJ. Development and comparative analysis of human urine exosome isolation strategies. Process Biochem. 2020;88:197–203. doi:10.1016/j.procbio.2019.09.017
54. Liu C, Guo JY, Tian F, et al. Field-free isolation of exosomes from extracellular vesicles by microfluidic viscoelastic flows. ACS Nano. 2017;11(7):6968–6976. doi:10.1021/acsnano.7b02277
55. Weng YJ, Sui ZG, Shan YC, et al. Effective isolation of exosomes with polyethylene glycol from cell culture supernatant for in-depth proteome profiling. Analyst. 2016;141(15):4640–4646. doi:10.1039/C6AN00892E
56. Chen JC, Li PL, Zhang TY, et al. Review on strategies and technologies for exosome isolation and purification. Front Bioeng Biotechnol. 2022;9:811971. doi:10.3389/fbioe.2021.811971
57. Kurian TK, Banik S, Gopal D, et al. Elucidating methods for isolation and quantification of exosomes: a review. Mol Biotechnol. 2021;63(4):249–266. doi:10.1007/s12033-021-00300-3
58. Gurunathan S, Kang MH, Jeyaraj M, et al. Review of the isolation, characterization, biological function, and multifarious therapeutic approaches of exosomes. Cells. 2019;8(4):307. doi:10.3390/cells8040307
59. Ghosh S, Bachilo SM, Weisman RB. Advanced sorting of single-walled carbon nanotubes by nonlinear density-gradient ultracentrifugation. Nat Nanotechnol. 2010;5(6):443–450. doi:10.1038/nnano.2010.68
60. Jang M, Kim S, Jeong H, et al. Affinity-mediated sorting order reversal of single-walled carbon nanotubes in density gradient ultracentrifugation. Nanotechnology. 2016;27(41):41LT01. doi:10.1088/0957-4484/27/41/41LT01
61. Yuana Y, Levels J, Grootemaat A, et al. Co-isolation of extracellular vesicles and high-density lipoproteins using density gradient ultracentrifugation. J Extracell Vesicles. 2014;8:3.
62. Martins TS, Vaz M, Henriques AG. A review on comparative studies addressing exosome isolation methods from body fluids. Anal Bioanal Chem. 2023;415(7):1239–1263. doi:10.1007/s00216-022-04174-5
63. Liangsupree T, Multia E, Riekkola ML. Modern isolation and separation techniques for extracellular vesicles. J Chromatogr A. 2021;1636:461773. doi:10.1016/j.chroma.2020.461773
64. Altıntaş Ö, Saylan Y. Exploring the versatility of exosomes: a review on isolation, characterization, detection methods, and diverse applications. Anal Chem. 2023;95(44):16029–16048. doi:10.1021/acs.analchem.3c02224
65. Huang K, Garimella S, Clay-Gilmour A, et al. Comparison of human urinary exosomes isolated via ultracentrifugation alone versus ultracentrifugation followed by SEC column-purification. J Pers Med. 2022;12(3):340. doi:10.3390/jpm12030340
66. Zarovni N, Corrado A, Guazzi P, et al. Integrated isolation and quantitative analysis of exosome shuttled proteins and nucleic acids using immunocapture approaches. Methods. 2015;87:46–58. doi:10.1016/j.ymeth.2015.05.028
67. Sharma P, Ludwig S, Muller L, et al. Immunoaffinity-based isolation of melanoma cell-derived exosomes from plasma of patients with melanoma. J Extracell Vesicles. 2018;7(1):1435138. doi:10.1080/20013078.2018.1435138
68. Hong CS, Muller L, Boyiadzis M, et al. Isolation and characterization of CD34+ blast-derived exosomes in acute myeloid leukemia. PLoS One. 2014;9(8):e103310. doi:10.1371/journal.pone.0103310
69. Althomali RH, Alshahrani SH, Almajidi YQ, et al. Current trends in nanomaterials-based electrochemiluminescence aptasensors for the determination of antibiotic residues in foodstuffs: a comprehensive review. Crit Rev Anal Chem. 2023;22:1–17. doi:10.1080/10408347.2023.2238059
70. Kuang JJ, Fu ZB, Sun XZ, et al. A colorimetric aptasensor based on a hemin/EpCAM aptamer DNAzyme for sensitive exosome detection. Analyst. 2022;147(22):5054–5061. doi:10.1039/D2AN01410F
71. Li C, Wang HY, Wei R, et al. An excellent colorimetric aptasensor integrating multifunctional SNAs and TdT-induced dual signal amplification for rapid sensitive detection of exosomes. Sens Actuators B Chem. 2023;380:133361. doi:10.1016/j.snb.2023.133361
72. An Y, Jin TY, Zhu YY, et al. An ultrasensitive electrochemical aptasensor for the determination of tumor exosomes based on click chemistry. Biosens Bioelectron. 2019;142:111503. doi:10.1016/j.bios.2019.111503
73. Zhu L, Xu YF, We XY, et al. Coupling aptamer-based protein tagging with metabolic glycan labeling for in situ visualization and biological function study of exosomal protein-specific glycosylation. Angew Chem Int Ed Engl. 2021;60(33):18111–18115. doi:10.1002/anie.202103696
74. You QN, Zhuang LL, Chang ZM, et al. Hierarchical Au nanoarrays functionalized 2D Ti2CTx MXene membranes for the detection of exosomes isolated from human lung carcinoma cells. Biosens Bioelectron. 2022;216:114647. doi:10.1016/j.bios.2022.114647
75. Yu CX, Li L, Liu LH, et al. Research of exosome in bone metastasis through dual aptamer recognition based entropy-driven amplification. Anal Biochem. 2022;636:114433. doi:10.1016/j.ab.2021.114433
76. Li T, Duan R, Duan ZJ, et al. Fluorescence signal amplification strategies based on DNA nanotechnology for miRNA detection. Chem Res Chin Univ. 2019;31:4.
77. Boriachek K, Islam MN, Gopalan V, et al. Quantum dot-based sensitive detection of disease specific exosome in serum. Analyst. 2017;142(12):2211–2219. doi:10.1039/C7AN00672A
78. Vinduska V, Gallops CE, O’Connor R, et al. Exosomal surface protein detection with quantum dots and immunomagnetic capture for cancer detection. Nanomaterials. 2021;11(7):1853. doi:10.3390/nano11071853
79. Li K, Tu JY, Zhang YL, et al. Ultrasensitive detection of exosomal miRNA with PMO-graphene quantum dots-functionalized field-effect transistor biosensor. iScience. 2022;25(7):104522. doi:10.1016/j.isci.2022.104522
80. Gao ML, Yin BC, Ye BC. Construction of a DNA-AuNP-based satellite network for exosome analysis. Analyst. 2019;144(20):5996–6003. doi:10.1039/C9AN01328H
81. Wang QL, Huang WX, Zhang PJ, et al. Colorimetric determination of the early biomarker hypoxia-inducible factor-1 alpha (HIF-1α) in circulating exosomes by using a gold seed-coated with aptamer-functionalized Au@Au core-shell peroxidase mimic. Microchim Acta. 2019;187(1):61. doi:10.1007/s00604-019-4035-z
82. Cheng WT, Duan CJ, Chen Y, et al. Highly sensitive aptasensor for detecting cancerous exosomes based on clover-like gold nanoclusters. Anal Chem. 2023;95(7):3606–3612. doi:10.1021/acs.analchem.2c04280
83. Huang WG, Yu YR, Yang CY, et al. Aptamer decorated magnetic graphene oxide nanoparticles for effective capture of exosomes. Chem Eng J. 2022;431:133849. doi:10.1016/j.cej.2021.133849
84. Lee J, Lee JH, Mondal J, et al. Magnetofluoro-immunosensing platform based on binary nanoparticle-decorated graphene for detection of cancer cell-derived exosomes. Int J Mol Sci. 2022;23(17):9619. doi:10.3390/ijms23179619
85. Huang J, Yan HD, Ma MX, et al. Hydroxylated graphene porous membrane-based biosensor for exosome isolation and detection. ACS Appl Nano Mater. 2022;5(5):6115–6124. doi:10.1021/acsanm.1c04336
86. Qi D, Zhang HW, Zhou ZY, et al. Preparation of CdTe quantum dots for detecting Cu (II) ions. Optical Materials. 2023;142:114048. doi:10.1016/j.optmat.2023.114048
87. Sun ZW, Li J, Yang YF, et al. Ratiometric fluorescent biosensor based on self-assembled fluorescent gold nanoparticles and duplex-specific nuclease-assisted signal amplification for sensitive detection of exosomal miRNA. Bioconjug Chem. 2022;33(9):1698–1706. doi:10.1021/acs.bioconjchem.2c00309
88. Chen SL, Chen CY, Hsun Hsieh JC, et al. Graphene oxide-based biosensors for liquid biopsies in cancer diagnosis. Nanomaterials. 2019;9(12):1725. doi:10.3390/nano9121725
89. Mo LT, He WQ, Li ZY, et al. Recent progress in the development of DNA-based biosensors integrated with hybridization chain reaction or catalytic hairpin assembly. Front Chem. 2023;11:1134863. doi:10.3389/fchem.2023.1134863
90. Zhang Y, Zhang XH, Situ B, et al. Rapid electrochemical biosensor for sensitive profiling of exosomal microRNA based on multifunctional DNA tetrahedron assisted catalytic hairpin assembly. Biosens Bioelectron. 2021;183:113205. doi:10.1016/j.bios.2021.113205
91. Zhou JQ, Lin QY, Huang ZP, et al. Aptamer-initiated catalytic hairpin assembly fluorescence assay for universal, sensitive exosome detection. Anal Chem. 2022;94:5723–5728.
92. Zhang LM, Gao QX, Chen J, et al. A universal catalytic hairpin assembly system for direct plasma biopsy of exosomal PIWI-interacting RNAs and microRNAs. Anal Chim Acta. 2022;1192:339382. doi:10.1016/j.aca.2021.339382
93. Wu TT, Liu XS, Chen HJ, et al. An in situ exosomal miRNA sensing biochip based on multi-branched localized catalytic hairpin assembly and photonic crystals. Biosens Bioelectron. 2023;222:115013. doi:10.1016/j.bios.2022.115013
94. Zhang YZ, Wang D, Yue S, et al. Sensitive multicolor visual detection of exosomes via dual signal amplification strategy of enzyme-catalyzed metallization of Au nanorods and hybridization chain reaction. ACS Sens. 2019;4(12):3210–3218. doi:10.1021/acssensors.9b01644
95. Zhang WF, Tian ZH, Yang SJ, et al. Electrochemical micro-aptasensors for exosome detection based on hybridization chain reaction amplification. Microsyst Nanoeng. 2021;7(1):63. doi:10.1038/s41378-021-00293-8
96. Kim J, Shim JS, Han BH, et al. Hydrogel-based hybridization chain reaction (HCR) for detection of urinary exosomal miRNAs as a diagnostic tool of prostate cancer. Biosens Bioelectron. 2021;192:113504. doi:10.1016/j.bios.2021.113504
97. Kang SY, Zhu L, Wang WC, et al. Amplified visualization and function exploration of exosomal protein-specific glycosylation using hybridization chain reaction from non-functional epitope. Sci China Chem. 2022;65(6):1204–1211. doi:10.1007/s11426-022-1240-5
98. Sun X, Chen J, Lang JL. Sensitive detection of exosomal miRNA for cardiovascular diseases with target initiate proximity ligation assay (TIPLA). Microchem J. 2020;158:105193. doi:10.1016/j.microc.2020.105193
99. Wang ZL, Zong SF, Liu Y, et al. Simultaneous detection of multiple exosomal microRNAs for exosome screening based on rolling circle amplification. Nanotechnology. 2021;32(8):085504. doi:10.1088/1361-6528/abc7d4
100. Li C, Zhou MY, Wang HY, et al. Rolling circle amplification assisted dual signal amplification colorimetric biosensor for ultrasensitive detection of leukemia-derived exosomes. Talanta. 2022;245:123444. doi:10.1016/j.talanta.2022.123444
101. Chen XL, Huang CW, Nie FP, et al. Enzyme-free and sensitive method for single-stranded nucleic acid detection based on CHA and HCR. Anal Methods. 2023;15(34):4243–4251. doi:10.1039/D3AY00975K
102. Xia YQ, Lei X, Ma XC, et al. Combination of RCA and DNAzyme for dual-signal isothermal amplification of exosome RNA. Molecules. 2023;28(14):5528. doi:10.3390/molecules28145528
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
