Back to Journals » International Journal of Nanomedicine » Volume 16
Exosomal lncRNA CHL1-AS1 Derived from Peritoneal Macrophages Promotes the Progression of Endometriosis via the miR-610/MDM2 Axis
Authors Liu T , Liu M, Zheng C, Zhang D, Li M, Zhang L
Received 10 June 2021
Accepted for publication 3 August 2021
Published 12 August 2021 Volume 2021:16 Pages 5451—5464
DOI https://doi.org/10.2147/IJN.S323671
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Ebrahim Mostafavi
Ting Liu,1 Mei Liu,2 Caihua Zheng,3 Daoyan Zhang,4 Mingbao Li,1 Lu Zhang1
1Department of Obstetrics and Gynecology, Qilu Hospital of Shandong University, Jinan, 250012, People’s Republic of China; 2Department of Obstetrics, Affiliated Hospital of Shandong University of Traditional Chinese Medicine, Jinan, 250011, People’s Republic of China; 3Department of Obstetrics, Changle County Hospital of Traditional Chinese Medicine, Changle, 262400, People’s Republic of China; 4Department of Obstetrics and Gynecology, People’s Hospital of Qihe County, Qihe, 251100, People’s Republic of China
Correspondence: Lu Zhang
Department of Obstetrics and Gynecology, Qilu Hospital of Shandong University, No. 107 Wenhua West Road, Jinan, Shandong Province, 250012, People’s Republic of China
Email [email protected]
Background: Exosomes secreted by peritoneal macrophages (pM&phis;) are deeply involved in the development of endometriosis (EMs). Exosomes can mediate cell-to-cell communication by transferring biological molecules. This study aimed to explore the effect and mechanism of exosomal long non-coding RNA (lncRNA) CHL1-AS1 derived from pM&phis; on EMs.
Materials and Methods: Exosomes (exo) from pM&phis; were isolated, identified, and co-cultured with ectopic endometrial stromal cells (eESCs) to investigate the biological functions of pM&phis;-exo. qRT-PCR was used to detect the expression of lncRNA CHL1-AS1 in pM&phis;-exo from EMs and control patients and verify the transportation of lncRNA CHL1-AS1 from pM&phis; to eESCs. The effects of exosomal lncRNA CHL1-AS1 on eESC proliferation, migration, invasion, and apoptosis were also detected. The relationships among lncRNA CHL1-AS1, miR-610, and MDM2 (mouse double minute 2) were verified by dual-luciferase reporter assay. The in vivo experiments were conducted to verify the effects of exosomal lncRNA on EMs using a xenograft model of EMs.
Results: Exosomes from pM&phis; were successfully isolated. EMs-pM&phis;-exo promoted eESC proliferation, migration, and invasion and inhibited their apoptosis. lncRNA CHL1-AS1 was upregulated in EMs-pM&phis;-exo and transported from pM&phis; to eESCs via exosomes. lncRNA CHL1-AS1 was found to act as a competing endogenous RNA of miR‑610 to promote the expression of MDM2. EMs-pM&phis;-exo shuttled lncRNA CHL1-AS1 to promote eESC proliferation, migration, and invasion and inhibit apoptosis by downregulating miR-610 and upregulating MDM2. Furthermore, exosomal lncRNA CHL1-AS1 promoted EMs lesions growth by increasing MDM2 in vivo.
Conclusion: The results demonstrate that exosomal lncRNA CHL1-AS1 promotes the proliferation, migration, and invasion of eESCs and inhibits their apoptosis by downregulating miR-610 and upregulating MDM2, which might be a potential therapeutic target for EMs.
Keywords: endometriosis, peritoneal macrophages, exosomes, lncRNA CHL1-AS1, miR-610, MDM2
Introduction
Endometriosis (EMs) is characterized by the persistence and growth of endometrial-like tissue outside the uterine cavity and especially in the pelvic cavity, which leads to pelvic pain, dysmenorrhea, dyspareunia, and infertility.1 EMs is present in 5–10% of reproductive-age women and 50% of infertile women.2 The management of EMs requires a multidisciplinary approach including endocrine therapy, surgery, and lifestyle interventions, but approximately 50% of patients with EMs have recurrent symptoms over 5 years, regardless of the proper treatment.3 Some possible reasons for EMs, including hormonal factors, genetics, and environmental factors, have been elucidated, but the exact causes of EMs remain unclear.4 Many studies have demonstrated that immune factors in the local environment contribute to the formation and progression of EMs.5 Dysfunction of various types of immune cells is integral to EMs pathogenesis.6 A recent report confirmed that peritoneal macrophages (pMφ) are deeply involved in EMs.7 Netrin-1 derived from pMφ can promote neuroangiogenesis in EMs.8 Indoleamine-pyrrole 2,3-dioxygenase-educated macrophages promoted the growth of endometrial stromal cells in EMs.9 Previous investigations only focused on the role of pMφ-derived cytokines in EMs, and other communications between pMφ and EMs lesions need further study.
Exosomes are lipid bilayer nanovesicles of 50–200 nm in diameter, which are secreted by almost all cells and can be found in a variety of body fluids.10 They mediate cell-to-cell communication by transferring biological molecules such as lipids, proteins, and non-coding RNAs.11 Recent studies have shown that macrophage-derived exosomes play a significant role in cancers,12 wound healing,13 and brain inflammation.14 Our previous research demonstrated that pMφ can affect the proliferation, migration, and invasion of ectopic endometrial stromal cells (eESCs) via the exosomal miR-22-3p/SIRT1/NF-kappa B pathway.15 However, the roles of other biomolecules in pMφ-derived exosomes (pMφ-exo) in EMs progression remain to be elucidated.
Long non-coding RNA (lncRNA) is a subgroup of non-coding RNAs with more than 200 nucleotides that can regulate gene expression by interacting with miRNAs, mRNAs, or proteins.16 LncRNAs packaged in exosomes are reportedly involved in mediating cell proliferation, migration, apoptosis, and autophagy.17 Exosomal lncRNA ROR1-AS1 accelerated the progression of glioma by regulating miR-4686.18 Many studies have reported that several lncRNAs are implicated in EMs,19 but the function of exosomal lncRNAs in EMs pathogenesis remains unknown. LncRNA CHL1-AS1 is a newly discovered lncRNA involved in EMs pathophysiology that may serve as a promoting factor.20 We hypothesized that transportation of exosomes from pMφ to eESCs upregulates the expression of lncRNA CHL1-AS1 in EMs, which acts as a miRNA sponge to regulate downstream target expression. The main objective of this study was to investigate the effect of pMφ-derived exosomal lncRNA CHL1-AS1 on eESCs and clarify the underlying molecular mechanisms.
Materials and Methods
Clinical Samples
This study was approved by the Ethics Committee of Qilu Hospital of Shandong University (approval number KYLL-2020KS-232). Written informed consent was obtained from all patients before enrollment in compliance with the Declaration of Helsinki. Patients with ovarian EMs cysts diagnosed by laparoscopic surgery and histopathologic examination served as the EMs group (n = 50). The control group (n = 50) included patients who underwent laparoscopic surgery for fallopian tube obstruction. The exclusion criteria were malignancy, other benign ovarian cysts, severe pelvic inflammation, and polycystic ovarian syndrome. The mean ages of the EMs and control groups were 33 ± 2.1 and 32 ± 2.6 years, respectively. All patients were recruited in Qilu hospital of Shandong University from July 2019 to January 2020 and had not received preoperative hormonal therapy or taken any medicine for at least 3 months. Peritoneal fluid samples were obtained during laparoscopy. Ectopic endometrial tissues were obtained from ovarian EMs cysts.
Culture and Characterization of eESCs
eESCs were isolated from ectopic endometrial tissues that were minced into small pieces, washed with Dulbecco’s modified Eagle’s medium (DMEM, Gibco, USA), and digested in 0.5% collagenase II (Life Technologies, USA) for 1 h at 37°C. The dispersed cells were passed through a 100-μm filter to remove undigested debris. The eESCs were collected by centrifugation at 1500 rpm for 10 min, washed twice with phosphate-buffered saline (PBS), and cultured in DMEM supplemented with 10% fetal bovine serum (FBS, Gibco, USA) and 1% penicillin-streptomycin solution at 37°C with 5% CO2. Vimentin, a biomarker of eESCs, was detected with an antibody (BD Biosciences, USA) using flow cytometry (FCM).
Isolation and Identification of pMφ
Under sterile conditions and direct vision, peritoneal fluid without blood contamination was collected at the time of laparoscopy by aspiration from the Douglas pouch before any surgical manipulations. The volume of peritoneal fluid ranged between 3 and 5 mL. Samples were placed in a sterile tube and centrifuged for 10 min at 400 × g to isolate pMφ according to previously described procedures.21 Briefly, the cell pellet was resuspended with the RPMI 1640 medium (Invitrogen, USA). Then, the cells were cultured in RPMI 1640 medium containing 10% FBS without exosomes in a humidified atmosphere of 5% CO2 at 37°C for 20 h. Non-adherent cells were discarded. Adherent cells were pMφ, which were used for subsequent experiments. To confirm the identity of pMφ, FCM was performed using the macrophage markers CD11b and CD68.
pMφ-Exo Isolation
The culture supernatants of pMφ were collected for exosome isolation by sequential centrifugation (Optima ultracentrifuge, Beckman Coulter, USA). The supernatants were centrifuged at 300 × g for 10 min followed by 2000 × g for 10 min to discard cells. Then, the supernatants were centrifuged at 10,000 × g for 30 min to discard cell debris. The supernatants were harvested and ultracentrifugated two times at 100,000 × g for 70 min each. The final pellets were exosomes, which were resuspended in 200 μL PBS and stored at −80°C.
Transmission Electron Microscope (TEM)
TEM was used to assess the morphology of exosomes. The exosomes were fixed in 2% paraformaldehyde solution (Solarbio, China), placed on copper grids, and allowed to stand for 10 min at room temperature. The copper grids were stained with 2% phosphotungstic acid solution (Solarbio, China). After drying for 10 min, the samples were observed and photographed under TEM (JEOL, Japan).
Nanoparticle-Tracking Analysis (NTA)
The size and concentration of exosomes were analyzed by NTA (NanoSight NS300, Malvern, UK). Exosomes suspensions were melted at 4°C and diluted with PBS (1:200), and the diluted exosomes suspension was injected into the sample chamber according to the manufacturer’s instructions. The Brownian motion of exosomes was observed and analyzed to calculate the size and concentration of exosomes using NTA software.
Western Blot Analysis
The RIPA lysate containing phenylmethanesulfonyl fluoride (Beyotime, China) was added into the exosomes or treated cells, followed by centrifuged at 10,000 × g for 20 min. The total protein concentration was determined by BCA kit (Thermo Fisher Scientific, USA). Each sample was separated by 10% separation gel SDS-PAGE electrophoresis, subsequently transferred onto the PVDF membranes, and blocked with 5% skim milk powder for 1 h at room temperature. The PVDF membranes were then incubated with the primary antibodies, including anti-CD9 (ab2215, 1:1000), anti-CD63 (ab59479, 1:1000), and anti-MDM2 (ab16895, 1:500) at 4°C overnight. The following day, the membrane was incubated with horseradish peroxidase-conjugated secondary antibody (ab6721, 1:2000) for 2 h at room temperature. Blots were detected by enhanced chemiluminescence. β-actin (ab18226, 1:1000) and GAPDH (ab181602; 1:10,000) were used as internal controls. All antibodies were purchased from Abcam Inc. UK.
Internalization of DiI-Labeled Exosomes
To determine whether pMφ-exo can be internalized by eESCs, 1μM DiI lipophilic dye (Invitrogen, USA) was used to label exosomes. After incubating at 37°C for 30 min, DiI-labeled exosomes were added to the culture medium of eESCs for 24 h. The treated eESCs were incubated with 4ʹ, 6-diamidino-2-phenylindole (DAPI, Invitrogen, USA) for 5 min at room temperature, and then viewed under a fluorescence microscope (Olympus, Japan).
Cell Counting Kit-8 (CCK-8) Assay
CCK-8 assay (Dojindo, Japan) was performed to assess cell viability according to the manufacturer’s protocol. Briefly, treated eESCs were plated on 96-well plates with 5×103 cells per well. Then, 10 μL of CCK-8 solution was added into each well followed by incubation at 37°C for 2 h. The optical density (OD) value at a wavelength of 450 nm was detected by a microplate reader (Thermo Fisher Scientific, USA) every 24 h for 5 days.
5-Ethynyl-2′-Deoxyuridine (EdU) Assay
Treated eESCs were plated on 96-well plates with 5×103 cells per well. Then cells were incubated with 100 μL 50 μm EdU solution (RiboBio, China) for 2 h, fixed with 50 μL 4% paraformaldehyde for 30 min, and finally incubated with 50 μL 2 mg/mL glycine for 5 min. Subsequently, the cells were incubated with 100 μL 0.5% Triton X-100 osmotic agent for 10 min and incubated in the dark with 100 μL Apollo solution for 30 min at room temperature, then infiltrated and decolorized with methanol. Finally, the cells were stained with DAPI and observed under a fluorescence microscope (Olympus, Japan).
Wound Healing Assay
eESCs were seeded into 24-well plates and incubated until they reached 90–100% confluence. Then the cell layer was scratched with a sterile plastic micropipette tip in the middle of each well, and loose cells were washed away with PBS. Wound healing was observed and photographed under an inverted microscope (Olympus, Japan) at 0 and 24 h after scratching.
Transwell Assays
For migration assay, eESCs suspended in 200 μL serum-free medium were seeded into the upper chambers. Complete medium containing FBS was placed into the lower chamber. After 24 h cultivation, cells in the lower chamber were washed with PBS, fixed with polyformaldehyde for 10 min, and stained with 0.1% crystal violet for 5 min. The migrated cells were observed and counted using an inverted microscope (Olympus, Japan). For invasion assay, eESCs were seeded into the upper chambers coated with Matrigel; the other steps were the same as above.
Flow Cytometry (FCM)
FCM was used to quantify the apoptosis of eESCs with an annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) staining assay kit (Solarbio, China) according to the manufacturer’s instructions. Briefly, eESCs (4 × 105) were washed twice with PBS and resuspended in the binding buffer. Then, 5 µL Annexin V-FITC and 5 µL PI were mixed with the cells and incubated for 15 min in the dark at room temperature. Finally, each sample was analyzed using a FACScan flow cytometer (BD Biosciences, USA).
Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
The total RNA was extracted using TRIzol reagent (Invitrogen, USA) and reversely transcribed into cDNA using an ImProm-II reverse transcription kit (Promega, USA) according to the manufacturer’s instructions. The cDNA samples were used for qRT-PCR with SYBR Primer-Script RT-PCR kits (Takara, Japan). U6 was used as the internal control for miR-610, while GAPDH served as the internal control for CHL1-AS1 and MDM2. The relative expression levels of RNA were calculated by using 2−ΔΔCt method. The experiment was repeated three times. The primers for CHL1-AS1, miR-610, MDM2, U6, and GAPDH are shown in Table 1.
 |
Table 1 The Primer Sequences Used for qRT-PCR |
CHL1-AS1-Interfering Exosomes
EMs-pMφ was transiently transfected with the small interfering RNA (siRNA) of CHL1-AS1 by LipofectamineTM 2000 reagent (Invitrogen, USA) for 48 h according to the manufacturer’s instructions, which was named as si-CHL1-AS1. EMs-pMφ-exosi-CHL1−AS1 and EMs-pMφ-exosi-control were isolated from CHL1-AS1-interfering or control-interfering EMs-pMφ based on the previous method of pMφ-exo isolation.
Cell Transfection
Cells were transiently transfected with CHL1-AS1 overexpressing vector, si-CHL1-AS1, miR-610 mimics, miR-610 inhibitors, or MDM2 overexpressing vector with LipofectamineTM 2000 reagent (Invitrogen, USA). The CHL1-AS1 overexpressing vector and its control, si-CHL1-AS1 and si-control, miR-610 mimics and its control (mimics NC), miR-610 inhibitors and its control (inhibitors NC), MDM2 overexpressing vector and its control were purchased from GeneChem (China).
Dual-Luciferase Reporter Gene Assay
CHL1-AS1 fragments and the 3ʹ-untranslated region (3ʹ-UTR) of MDM2 containing the predicted wild-type (wt) binding sites of miR-610 or mutated miR-610 binding sites (mut) were amplified by PCR and inserted into a luciferase reporter vector. These constructs were named as CHL1-AS1-wt, CHL1-AS1-mut, MDM2-wt, and MDM2-mut, which were transfected into eESCs with LipofectamineTM 2000 (Invitrogen, USA). After 24 h, luciferase activity was analyzed by a dual luciferase reporter assay system (Promega, USA).
RNA Immunoprecipitation (RIP) Assay
The RIP kit (Millipore, USA) was used to detect the binding of lncRNA CHL1-AS1 to Ago2. eESCs were washed twice with PBS. After adding RIPA lysis buffer (Beyotime, China), the cells were lysed on ice for 30 min and centrifuged at 14,000 × g for 10 min at 4°C. The supernatant was collected. A subset of the cell extract was used as an input, and the rest was incubated with Ago2 antibody (Abcam, UK) for co-precipitation. The magnetic bead-antibody complex was resuspended in 900 μL RIP wash buffer and incubated at 4°C with 100 μL cell extract. The sample was placed on a magnetic pedestal to collect the magnetic bead protein complex. The RNA was extracted from the sample and input after protease K detachment, followed by qRT-PCR. Anti-IgG (Abcam, UK) was used as the negative control.
RNA Pull-Down Assay
eESCs were lysed in a specific lysis buffer (Ambion, USA), and the lysates were incubated with M-280 streptavidin beads (Sigma-Aldrich, USA) that were precoated with RNase-free bovine serum albumin and yeast tRNA (Sigma-Aldrich, USA). The beads were incubated at 4°C for 3 h, washed twice with pre-cooled pyrolysis buffer, three times with low-salt buffer, and once with high-salt buffer. The bound RNA was purified by TRIzol, and the enrichment of CHL1-AS1 was verified by qRT-PCR.
Establishment of the EMs Mouse Model
The mouse model of EMs was established as previously described.22 In brief, the mice were anesthetized with 4% chloral hydrate (4 g/100 mL) and fixed an operating surface. A 2.0-cm segment of the uterine horn was excised via a vertical incision on the abdomen and placed in saline solution. The endometrium was carefully extracted from muscles and trimmed into two 5 mm × 5 mm pieces. A subcutaneous pocket was fashioned on each side of the abdominal wall. The uterine segments were then placed in the space where the endometrium faced the abdominal muscles. Six-week-old female nude mice were purchased from Beijing HFK Bioscience Company (China). The EMs mice were divided into four groups: PBS, Con-pMφ-exo, EMs-pMφ-exo, and EMs-pMφ-exosi-CHL1-AS1. All groups received the treatments via tail vein injections that were repeated every 2 days until Day 14. Mice were sacrificed 24 h after the last injection, and EMs lesions were collected for further experiments. The weight of lesions was recorded, and the size of lesions was assessed with the formula of volume = π/6 × length × width × height. The animal experiments were approved by the Ethics Committee of Qilu Hospital of Shandong University and conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals of the National Institutes of Health.
Statistical Analysis
All data were analyzed by GraphPad 7.0 software (GraphPad Software, USA). The measurement data were expressed as mean ± standard deviation. Comparisons between two groups were assessed with Student’s t-tests. One-way analysis of variance (ANOVA) was utilized to evaluate the difference between multiple groups followed by Tukey’s post hoc test. All experiments were repeated at least 3 times. P value less than 0.05 was considered statistically significant.
Results
Identification of eESCs, pMφ, and pMφ-Exo
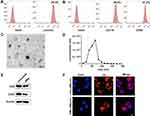
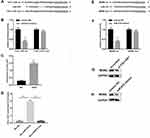
Data in previous study suggested that vimentin was a biomarker for eESCs, while positive biomarkers for pMφ included CD11b and CD68. In this study, FCM confirmed positive vimentin expression in eESCs (Figure 1A) and positive CD11b and CD68 expression in pMφ (Figure 1B). As shown in Figure 1C, the morphology of pMφ-exo under TEM was a round or oval shape. NTA demonstrated that the diameter of exosomes ranged from 50 to 200 nm, and the mean diameter was 118 ± 5.7nm (Figure 1D). Western blot revealed that pMφ-exo highly expressed the exosomal markers CD9 and CD63 (Figure 1E). After DiI-labeled pMφ-exo were incubated with DAPI-labeled eESCs for 24 h, fluorescence images showed that red pMφ-exo localized on the cytoplasm of eESCs, which indicated that eESC uptake of DiI-labeled exosomes (Figure 1F).
EMs-pMφ-Exo Promote eESC Proliferation, Migration, and Invasion and Inhibit Their Apoptosis
To explore the effect of pMφ-exo on the activity of eESCs, eESCs were incubated with PBS, EMs-pMφ-exo, or Con-pMφ-exo. As shown in Figure 2A and B, eESC proliferation was obviously increased in the EMs-pMφ-exo group compared to the PBS and Con-pMφ-exo groups. Compared with PBS and Con-pMφ-exo, EMs-pMφ-exo significantly enhanced eESC migration and invasion (Figure 2C and D). Moreover, the addition of EMs-pMφ-exo significantly inhibited eESC apoptosis (Figure 2E). There was no significant difference in the effects of PBS and Con-pMφ-exo on eESCs.
EMs-pMφ-Exo Transports lncRNA CHL1-AS1 into eESCs
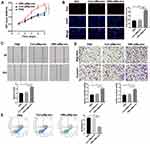
The expressions of lncRNA CHL1-AS1 were compared between EMs-pMφ-exo and Con-pMφ-exo by qRT-PCR, and the results showed an increase in the EMs-pMφ-exo group (Figure 3A). To confirm whether EMs-pMφ-exo can transport lncRNA CHL1-AS1 into eESCs, eESCs were co-cultured with EMs-pMφ-exo or Con-pMφ-exo for 48 h. Treatment with EMs-pMφ-exo induced an increase of lncRNA CHL1-AS1 in eESCs compared with Con-pMφ-exo treatment (Figure 3B). Furthermore, we interfered with the expression of lncRNA CHL1-AS1 in EMs-pMφ-exo by transfecting EMs-pMφ with si-CHL1-AS1 or si-control. Then EMs-pMφ-exosi-CHL1-AS1 and EMs-pMφ-exosi-control were isolated from treated EMs-pMφ (Figure 3C). The expression of lncRNA CHL1-AS1 in eESCs was reduced after co-culturing with EMs-pMφ-exosi-CHL1-AS1 (Figure 3D). This evidence strongly suggested that pMφ can affect lncRNA CHL1-AS1 expression in eESCs through the exosomal pathway.
Exosomal lncRNA CHL1-AS1 from EMs-pMφ Enhances the Proliferation, Migration, and Invasion of eESCs and Represses Their Apoptosis
Given the effect of EMs-pMφ-exo on eESCs and the increase of lncRNA CHL1-AS1 in EMs-pMφ-exo, we explored the effect of exosomal lncRNA CHL1-AS1 on eESCs. After co-culturing with Con-pMφ-exo, EMs-pMφ-exo, EMs-pMφ-exosi-control, or EMs-pMφ-exosi-CHL1-AS1 we measured the proliferation, migration, invasion, and apoptosis of eESCs with CCK-8, EdU assay, wound healing, Transwell assays, and FCM, respectively. As shown in Figure 4A and B, the proliferation of eESCs incubated with EMs-pMφ-exosi-CHL1-AS1 was clearly attenuated compared to those incubated with EMs-pMφ-exo and EMs-pMφ-exosi-control. eESC migration and invasion were blocked in the EMs-pMφ-exosi-CHL1-AS1 group compared with the EMs-pMφ-exo and EMs-pMφ-exosi-control groups (Figure 4C and D). Finally, FCM revealed that compared with EMs-pMφ-exo and EMs-pMφ-exosi-control, EMs-pMφ-exosi-CHL1-AS1 significantly promoted eESC apoptosis (Figure 4E).
LncRNA CHL1-AS1 Acts as a Competing Endogenous RNA (ceRNA) by Sponging miR-610 to Regulate MDM2
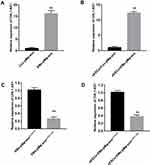
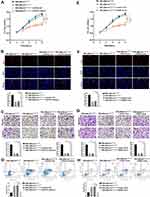
The DIANA tools online database predicted potential binding sites between lncRNA CHL1-AS1 and miR-610 (Figure 5A). The dual-luciferase reporter assay showed that the co-transfection with miR-610 mimics and CHL1-AS1-wt vector significantly lowered the luciferase activity, while co-transfection with miR-610 mimics and CHL1-AS1-mut vector did not affect the luciferase activity (Figure 5B). RIP assay revealed that there was a higher specific adsorption level of lncRNA CHL1-AS1 on Ago2 (Figure 5C), and the RNA pull-down results revealed that the enrichment level of lncRNA CHL1-AS1 was increased in the Bio-miR-610-wt group compared to the Bio-miR-610-mut group (Figure 5D). These findings indicated that CHL1-AS1 and miR-610 could bind to each other.
We also predicted the target genes of miR-610 on the Targetscan website and found binding sites between miR-610 and MDM2 (Figure 5E). Dual-luciferase reporter assay demonstrated suppressed luciferase activity in the miR-610 mimics + MDM2-wt group compared with the miR-610 mimics + MDM2-mut group (Figure 5F). Furthermore, eESCs treated with si-CHL1-AS1 or miR-610 mimics showed reduced protein expression of MDM2 (Figure 5G and H).
EMs-pMφ-Exo Shuttled lncRNA CHL1-AS1 Downregulates miR-610 and Upregulates MDM2 to Promote the Proliferation, Migration, and Invasion of eESCs and Inhibit Their Apoptosis
We further explored whether lncRNA CHL1-AS1 affected eESC activity by regulating the miR-610/MDM2 axis. The results of CCK-8 and EdU assay showed that silencing miR-610 could reverse the inhibitory effects of EMs-pMφ-exosi-CHL1-AS1 on eESC proliferation (Figure 6A and B). Notably, miR-610 inhibitors abrogated the inhibitory effects of EMs-pMφ-exosi-CHL1-AS1 on eESC migration and invasion (Figure 6C). Silencing miR-610 diminished the apoptosis-promoting effect of EMs-pMφ-exosi-CHL1-AS1 (Figure 6D).
To examine the role of MDM2 in the effect of lncRNA CHL1-AS1 on eESCs, MDM2 was upregulated by transfection with an MDM2 overexpression vector. The results showed that MDM2 overexpression reversed the suppressive influence of EMs-pMφ-exosi-CHL1-AS1 on eESC proliferation, migration, and invasion (Figure 6E–G). Moreover, MDM2 overexpression diminished the apoptosis-promoting effect of EMs-pMφ-exosi-CHL1-AS1 (Figure 6H).
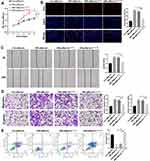
LncRNA CHL1-AS1 in EMs-pMφ-Exo Promotes EMs Lesions Growth by Increasing MDM2 in vivo
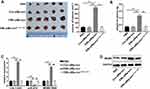
An EMs mouse model was established to elucidate the effect of exosomal lncRNA CHL1-AS1 derived from EMs-pMφ on EMs lesions growth in vivo. Compared with the PBS and Con-pMφ-exo groups, the volume and weight of EMs lesions were increased in the EMs-pMφ-exo group, but EMs-pMφ-exosi-CHL1-AS1 injection reduced the volume and weight of EMs lesions compared with EMs-pMφ-exo injection (Figure 7A and B). To verify whether these effects were due to the abnormal expression of lncRNA CHL1-AS1, miR-610, and MDM2, we performed qRT-PCR and Western blot. As shown in Figure 7C, the expression of CHL1-AS1 and MDM2 mRNA was enhanced by EMs-pMφ-exo injection and reduced by EMs-pMφ-exosi-CHL1-AS1 injection, while the changes in the expression of miR-610 showed a reverse phenomenon. MDM2 protein expression was enhanced and reduced by EMs-pMφ-exo and EMs-pMφ-exosi-CHL1-AS1 injection, respectively (Figure 7D).
Discussion
Reciprocal communication between pMφ and eESCs is conducive to the establishment of EMs.23 Exosomes are an important medium that play an important role in EMs development.24 In the present study, we detected the effect of pMφ-exo on eESCs and confirmed that EMs-pMφ-exo could promote their proliferation, migration, and invasion and inhibit their apoptosis. The results provide a new link between pMφ and eESCs.
As exosomal lncRNA plays an important role in cell-to-cell communications,25 we hypothesized that pMφ-exo might exert their effect on eESCs via lncRNA. Our results demonstrated that lncRNA CHL1-AS1 was highly expressed in EMs-pMφ-exo and could be transported from pMφ to eESCs via the exosomal pathway. Interestingly, EMs-pMφ-exo enhanced the proliferation, migration, and invasion and repressed the apoptosis of eESCs via lncRNA CHL1-AS1. Following lncRNA CHL1-AS1 knockdown in EMs-pMφ, we obtained EMs-pMφ-exosi-CHL1-AS1 that contain lower amounts of lncRNA CHL1-AS1 than normal EMs-pMφ-exo or EMs-pMφ-exosi-control, and the effect of EMs-pMφ-exosi-CHL1-AS1 on eESCs was repressed compared to the EMs-pMφ-exosi-control group. This indicated that the knockdown of lncRNA CHL1-AS1 in EMs-pMφ exerted a suppressive effect and may be a novel therapeutic target for EMs.
LncRNAs have been well known to exert effects by acting as a ceRNA to regulate the post-transcription of miRNAs.26 Using the DIANA tools online database, we noted that CHL1-AS1 contained putative binding sites of miR-610, which was identified by dual-luciferase reporter assay. Sun et al reported that miR-610 suppressed colorectal cancer cell proliferation and invasion by repressing hepatoma-derived growth factor.27 Wang et al demonstrated that miR-610 blocked gastric cancer cell migration and invasion by inhibiting the expression of vasodilator-stimulated phosphoprotein.28 Another study showed that miR-610 suppressed glioblastoma cell proliferation through direct suppression of CCND2 and AKT3 expression.29 In addition, lncRNA FEZF1-AS1 promoted the growth of multiple myeloma cells by downregulating miR-610.30 However, there are no reports of a relationship between miR-610 and lncRNA CHL1-AS1 in eESCs. We found that silencing miR-610 reversed the inhibitory effects of EMs-pMφ-exosi-CHL1-AS1 on eESC proliferation, and these findings demonstrated that exosomal lncRNA CHL1-AS1 exerts its effects on eESCs through sponging miR-610, and miR-610 overexpression inhibited EMs progression.
Moreover, we further explored the downstream mechanism of miR-610. The bioinformatics analysis showed that miR-610 interacted with the 3ʹ-UTR of MDM2, and dual-luciferase report assay revealed that miR-610 suppressed the expression of MDM2 at the post-transcriptional level. MDM2, which is located in segment 13–14 of the long arm of chromosome 12, is found in the double minute chromosome of the transformed murine cell line.31 Accumulating evidence indicates that MDM2 can enhance cellular activity and promote tumor growth,32,33 which could reveal a new therapeutic strategy for EMs. Previous studies indicated that the positivity rate of MDM2 expression in normal endometrium was lower than that in EMs,34,35 which suggests that MDM2 serves as a promoting factor in EMs. In this work, lncRNA CHL1-AS1 enhanced the expression of MDM2 in vivo and in vitro, and MDM2 overexpression reversed the suppressive influence of EMs-pMφ-exosi-CHL1-AS1 on eESC proliferation, migration, and invasion, which indicated that MDM2 promoted EMs progression. These results are consistent with previous studies. Moreover, we found that lncRNA CHL1-AS1 in EMs-pMφ-exo promoted EMs lesions growth in vivo by increasing MDM2.
Although we identified exosomal lncRNA CHL1-AS1 as a promoter of EMs and clarified its downstream mechanism, there are several limitations of our study. Firstly, current methods employed for exosome isolation include sequential centrifugation, ultrafiltration, precipitation, and immunoaffinity-based capture, but they pose a variety of challenges.36 We selected classic sequential centrifugation, which require long run times and may damage exosomal cargo. It would be helpful to explore a new method to improve exosome purity and reduce cargo loss. Secondly, this is the first evidence that miR-670 was involved in EMs pathology, which is inconsistent with previous studies using circulating miRNAs microarrays.37,38 This may be due to the heterogeneity of EMs, the use of different technological platforms, and test sample variability. Further investigation is required to confirm the expression and function of miR-670 in EMs.
Conclusion
In summary, our findings indicate that EMs-pMφ-exo promoted EMs development by transporting lncRNA CHL1-AS1 to eESCs. lncRNA CHL1-AS1 acted as a sponge for miR-610, which upregulated the expression of MDM2 in vivo and in vitro. This study provides a new insight into the mechanism by which pMφ-exo modulate EMs progression, and suggests novel therapeutic strategies for EMs.
Acknowledgments
This research was supported by the Natural Science Foundation of Shandong Province, china (ZR2020QH043, ZR2020QH249).
Disclosure
The authors report no conflicts of interest for this work.
References
1. Zondervan KT, Becker CM, Missmer SA. Endometriosis. N Engl J Med. 2020;382(13):1244–1256.
2. Parazzini F, Esposito G, Tozzi L, Noli S, Bianchi S. Epidemiology of endometriosis and its comorbidities. Eur J Obstet Gynecol Reprod Biol. 2017;209:3–7.
3. Becker CM, Gattrell WT, Gude K, Singh SS. Reevaluating response and failure of medical treatment of endometriosis: a systematic review. Fertil Steril. 2017;108(1):125–136.
4. Vercellini P, Vigano P, Somigliana E, Fedele L. Endometriosis: pathogenesis and treatment. Nat Rev Endocrinol. 2014;10(5):261–275.
5. Symons LK, Miller JE, Kay VR, et al. The immunopathophysiology of endometriosis. Trends Mol Med. 2018;24(9):748–762.
6. Izumi G, Koga K, Takamura M, et al. Involvement of immune cells in the pathogenesis of endometriosis. J Obstet Gynaecol Res. 2018;44(2):191–198.
7. Capobianco A, Rovere-Querini P. Endometriosis, a disease of the macrophage. Front Immunol. 2013;4:9.
8. Guo X, Ding S, Li T, et al. Macrophage-derived netrin-1 is critical for neuroangiogenesis in endometriosis. Int J Biol Macromol. 2020;148:226–237.
9. Mei J, Chang KK, Sun HX. Immunosuppressive macrophages induced by IDO1 promote the growth of endometrial stromal cells in endometriosis. Mol Med Rep. 2017;15(4):2255–2260.
10. Gurunathan S, Kang MH, Qasim M, Khan K, Kim JH. Biogenesis, membrane trafficking, functions, and next generation nanotherapeutics medicine of extracellular vesicles. Int J Nanomedicine. 2021;16:3357–3383.
11. Zhang Y, Bi J, Huang J, Tang Y, Du S, Li P. Exosome: a review of its classification, isolation techniques, storage, diagnostic and targeted therapy applications. Int J Nanomedicine. 2020;15:6917–6934.
12. Liu J, Wu F, Zhou H. Macrophage-derived exosomes in cancers: biogenesis, functions and therapeutic applications. Immunol Lett. 2020;227:102–108.
13. Li M, Wang T, Tian H, Wei G, Zhao L, Shi Y. Macrophage-derived exosomes accelerate wound healing through their anti-inflammation effects in a diabetic rat model. Artif Cells Nanomed Biotechnol. 2019;47(1):3793–3803.
14. Yuan D, Zhao Y, Banks WA, et al. Macrophage exosomes as natural nanocarriers for protein delivery to inflamed brain. Biomaterials. 2017;142:1–12.
15. Zhang L, Li HH, Yuan M, Li D, Wang GY. Exosomal miR-22-3p derived from peritoneal macrophages enhances proliferation, migration, and invasion of ectopic endometrial stromal cells through regulation of the SIRT1/NF-kappaB signaling pathway. Eur Rev Med Pharmacol Sci. 2020;24(2):571–580.
16. Yao RW, Wang Y, Chen LL. Cellular functions of long noncoding RNAs. Nat Cell Biol. 2019;21(5):542–551.
17. Zhao W, Liu Y, Zhang C, Duan C. Multiple roles of exosomal long noncoding RNAs in cancers. Biomed Res Int. 2019;2019:1460572.
18. Chai Y, Wu HT, Liang CD, You CY, Xie MX, Xiao SW. Exosomal lncRNA ROR1-AS1 derived from tumor cells promotes glioma progression via regulating miR-4686. Int J Nanomedicine. 2020;15:8863–8872.
19. Panir K, Schjenken JE, Robertson SA, Hull ML. Non-coding RNAs in endometriosis: a narrative review. Hum Reprod Update. 2018;24(4):497–515.
20. Zhang C, Wu W, Ye X, et al. Aberrant expression of CHL1 gene and long non-coding RNA CHL1-AS1, CHL1-AS2 in ovarian endometriosis. Eur J Obstet Gynecol Reprod Biol. 2019;236:177–182.
21. Khan KN, Masuzaki H, Fujishita A, et al. Regulation of hepatocyte growth factor by basal and stimulated macrophages in women with endometriosis. Hum Reprod. 2005;20(1):49–60.
22. Liang Z, Chen Y, Zhao Y, et al. miR-200c suppresses endometriosis by targeting MALAT1 in vitro and in vivo. Stem Cell Res Ther. 2017;8(1):251.
23. Eyster KM, Hansen KA, Winterton E, Klinkova O, Drappeau D, Mark-Kappeler CJ. Reciprocal communication between endometrial stromal cells and macrophages. Reprod Sci. 2010;17(9):809–822.
24. Shomali N, Hemmatzadeh M, Yousefzadeh Y, et al. Exosomes: emerging biomarkers and targets in folliculogenesis and endometriosis. J Reprod Immunol. 2020;142:103181.
25. Wang M, Zhou L, Yu F, Zhang Y, Li P, Wang K. The functional roles of exosomal long non-coding RNAs in cancer. Cell Mol Life Sci. 2019;76(11):2059–2076.
26. Chen YG, Satpathy AT, Chang HY. Gene regulation in the immune system by long noncoding RNAs. Nat Immunol. 2017;18(9):962–972.
27. Sun B, Gu X, Chen Z, Xiang J. MiR-610 inhibits cell proliferation and invasion in colorectal cancer by repressing hepatoma-derived growth factor. Am J Cancer Res. 2015;5(12):3635–3644.
28. Wang J, Zhang J, Wu J, et al. MicroRNA-610 inhibits the migration and invasion of gastric cancer cells by suppressing the expression of vasodilator-stimulated phosphoprotein. Eur J Cancer. 2012;48(12):1904–1913.
29. Mo X, Cao Q, Liang H, Liu J, Li H, Liu F. MicroRNA-610 suppresses the proliferation of human glioblastoma cells by repressing CCND2 and AKT3. Mol Med Rep. 2016;13(3):1961–1966.
30. Li QY, Chen L, Hu N, Zhao H. Long non-coding RNA FEZF1-AS1 promotes cell growth in multiple myeloma via miR-610/Akt3 axis. Biomed Pharmacother. 2018;103:1727–1732.
31. Cahilly-Snyder L, Yang-Feng T, Francke U, George DL. Molecular analysis and chromosomal mapping of amplified genes isolated from a transformed mouse 3T3 cell line. Somat Cell Mol Genet. 1987;13(3):235–244.
32. Wang W, Wang X, Rajaei M, et al. Targeting MDM2 for neuroblastoma therapy: in vitro and in vivo anticancer activity and mechanism of action. Cancers (Basel). 2020;12:12.
33. Khurana A, Shafer DA. MDM2 antagonists as a novel treatment option for acute myeloid leukemia: perspectives on the therapeutic potential of idasanutlin (RG7388). Onco Targets Ther. 2019;12:2903–2910.
34. Goumenou A, Panayiotides I, Mahutte NG, Matalliotakis I, Fragouli Y, Arici A. Immunohistochemical expression of p53, MDM2, and p21Waf1 oncoproteins in endometriomas but not adenomyosis. J Soc Gynecol Investig. 2005;12(4):263–266.
35. Sang L, Fang QJ, Zhao XB. A research on the protein expression of p53, p16, and MDM2 in endometriosis. Medicine (Baltimore). 2019;98(14):e14776.
36. Kurian TK, Banik S, Gopal D, Chakrabarti S, Mazumder N. Elucidating methods for isolation and quantification of exosomes: a review. Mol Biotechnol. 2021;63(4):249–266.
37. Gu CL, Zhang Z, Fan WS, et al. Identification of MicroRNAs as potential biomarkers in ovarian endometriosis. Reprod Sci. 2020;27(9):1715–1723.
38. Wang WT, Zhao YN, Han BW, Hong SJ, Chen YQ. Circulating microRNAs identified in a genome-wide serum microRNA expression analysis as noninvasive biomarkers for endometriosis. J Clin Endocrinol Metab. 2013;98(1):281–289.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.