Back to Journals » Cancer Management and Research » Volume 12
Exosomal Circ-ZNF652 Promotes Cell Proliferation, Migration, Invasion and Glycolysis in Hepatocellular Carcinoma via miR-29a-3p/GUCD1 Axis
Authors Li Y, Zang H, Zhang X, Huang G
Received 22 April 2020
Accepted for publication 30 July 2020
Published 24 August 2020 Volume 2020:12 Pages 7739—7751
DOI https://doi.org/10.2147/CMAR.S259424
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Yuhui Li, Hongliang Zang, Xue Zhang, Guomin Huang
Department of General Surgery, China-Japan Union Hospital of Jilin University, Changchun 130012, People’s Republic of China
Correspondence: Guomin Huang Tel +86-431-84997753
Email [email protected]
Background: Circular RNAs (circRNAs) play a crucial role in hepatocellular carcinoma (HCC) progression. However, the role of exosomal circRNAs in HCC is still largely unknown. We aimed to explore the function of exosomal circ-ZNF652 in HCC.
Methods: The morphology and size of exosomes were examined by transmission electron microscopy (TEM) and nanoparticle tracking analysis (NTA). The expression of circ-ZNF652, ZNF652 mRNA, microRNA-29a-3p (miR-29a-3p) and guanylyl cyclase domain containing 1 (GUCD1) mRNA was determined by quantitative real-time polymerase chain reaction (qRT-PCR). The protein levels of CD63, CD81, hexokinase 2 (HK2) and GUCD1 were examined via Western blot assay. The stability of circ-ZNF652 was examined by RNase R digestion assay. Cell proliferation was analyzed by 3-(4, 5-dimethyl-2-thiazolyl)-2, 5-diphenyl-2-H-tetrazolium bromide (MTT) assay. Cell migration and invasion were assessed by transwell assay. The glycolysis level was detected via specific kits. The association between miR-29a-3p and circ-ZNF652 or GUCD1 was analyzed by dual-luciferase reporter assay and RNA immunoprecipitation (RIP) assay. A murine xenograft model was constructed to explore the effect of circ-ZNF652 in vivo.
Results: Exosomal circ-ZNF652 was upregulated in HCC patients’ serums and HCC cells. Exosomal circ-ZNF652 could transfer to HCC cells, and circ-ZNF652 silencing suppressed HCC cell proliferation, migration, invasion and glycolysis. Circ-ZNF652 was a sponge of miR-29a-3p, and the inhibitory effect of circ-ZNF652 silencing on HCC cell progression was weakened by miR-29a-3p inhibitor. GUCD1 was a target gene of miR-29a-3p, and GUCD1 overexpression restored the effect of miR-29a-3p on HCC cell development. Moreover, circ-ZNF652 knockdown repressed tumor growth in vivo.
Conclusion: Exosomal circ-ZNF652 contributes to HCC cell proliferation, migration, invasion and glycolysis by miR-29a-3p/GUCD1 axis.
Keywords: HCC, exosome, circ-ZNF652, miR-29a-5p, GUCD1
Introduction
As a primary liver cancer, hepatocellular carcinoma (HCC) has shown high incidence and mortality.1 In the past decades, there is no doubt that great progress has been made in the diagnosis and therapy of HCC. However, the overall survival remains very dismal due to the high metastasis rate and high recurrence rate.2,3 Thus, it is urgent to explore the underlying mechanism of HCC and find novel effective therapeutic targets for patients with HCC.
Exosomes, small vesicles with a diameter of 50–140 nm that secreted by diverse cell types, have been verified to exert a critical effect on cancer progression.4,5 It has been documented that exosomes function as vital mediators in cell-cell communication via delivering proteins, circular RNAs (circRNAs), microRNAs (miRNAs), mRNAs or other bioactive substances.6 CircRNAs, a novel class of non-coding RNAs (ncRNAs) with closed-loop structures, take part in the regulation of multiple cancers, including HCC.7 For instance, circ-FOXP1 depletion hampered cell metastasis and growth and facilitated cell death in HCC.8 High expression of circ_0021093 was observed in HCC and associated with a poor prognosis.9 Circ-SETD3 repressed cell growth and induced cell cycle arrest in HCC.10 These findings indicated that circRNAs played dual roles in HCC development. Circ-ZNF652 has been demonstrated to act as a positive regulator in HCC progression.11 Recently, circRNAs were found to be enriched in exosomes and could be biomarkers in various diseases.12 However, the potential role of exosomal circ-ZNF652 is still unclear.
MiRNAs are a series of ncRNAs with 18–24 nucleotides and have essential effects on multiple biological processes.13 It is widely accepted that miRNAs can modulate gene expression via interacting with the 3ʹ-untranslated region (3ʹUTR) of target mRNAs.14 Numerous literature has exhibited that miRNAs can affect HCC development. For example, Chen et al disclosed that miR-361a-5p hampered HCC growth and metastasis by targeting WT1.15 Gao et al declared that miR-217 inhibited HCC development via interacting with KLF5.16 Xiao et al implicated that miR-29a-3p could bind to Mdm2 or PDGF-B to take part in the regulation of HCC growth.17 Guanylyl cyclase domain containing 1 (GUCD1) is a widely expressed and highly conserved gene and plays crucial roles in liver regeneration and tumorigenesis.18 However, whether miR-29a-3p can target GUCD1 to affect HCC development is still barely documented.
Here, we determined the expression of exosomal circ-ZNF652 in HCC patients’ serums and cells. Besides, we investigated the effects of circ-ZNF652 in HCC cell proliferation, metastasis and glycolysis. Mechanically, the underlying mechanism of circ-ZNF652 in HCC was explored.
Materials and Methods
Serum Sample Collection
Thirty-three serum samples were collected from HCC patients and healthy volunteers at China–Japan Union Hospital of Jilin University and saved at −80°C before use. The study obtained permission from the Ethics Committee of China–Japan Union Hospital of Jilin University. All participants were provided written informed consents prior to this experiment.
Cell Culture
HCC cell lines SNU-387 and Huh7 were bought from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Normal liver cell line THLE-2 was bought from the American Type Culture Collection (ATCC, Manassas, VA, USA). All cells were grown in RPMI 1640 medium (Gibco, Grand Island, NY, USA) including 10% fetal bovine serum (FBS; Gibco) and 1% penicillin-streptomycin (Gibco) at 37°C in an incubator with 5% CO2.
Exosome Isolation
The exosomes were isolated from serum and cell culture medium with ExoQuick precipitation kit (System Biosciences, Mountain View, CA, USA). In brief, serums or cell culture medium was collected and centrifuged for 30 min at 3000×g to remove dead cells and cellular debris. Then, ExoQuick solution was added into the supernatant and incubated at 4°C for 30 min. After being blended, the mix was centrifuged at 1500×g for 30 min. Next, the supernatant was discarded followed by centrifugation for 5 min at 1500×g to remove the extra liquids. Exosome pellets were resuspended in phosphate-buffered saline (PBS; Solarbio, Beijing, China) and saved at −80°C.
Transmission Electron Microscopy (TEM)
Exosomes were put on a carbon-coated copper grid and incubated for 3 min, stained with phosphotungstic acid solution and dried at 65°C. The morphology of exosomes was determined using an EM-1200EX TEM (JEOL Ltd., Tokyo, Japan).
Nanoparticle Tracking Analysis (NTA)
Zetasizer (Malvern Panalytical Ltd., Malvern, UK, USA) was utilized for the detection of the size distribution of exosomes based on the instructions of the manufacturer.
Western Blot Assay
Serums, cells or exosomes were lysed in RIPA buffer (Beyotime, Shanghai, China) to extract total protein. After being determined with a BCA protein assay kit (Tiangen, Beijing, China), the proteins were separated via sodium dodecyl sulfonate-polyacrylamide gel (SDS-PAGE; Solarbio). Then, the proteins were transferred onto polyvinylidene difluoride membranes (PVDF; Pall Corporation, New York, NYC, USA) and blocked in skim milk for 2 h. Afterward, the membranes were incubated with primary antibodies: CD63 (ab216130; Abcam, Cambridge, MA, USA), CD81 (ab109201; Abcam), hexokinase 2 (HK2; ab209847; Abcam), GUCD1 (ab224743; Abcam) or GAPDH (ab9485; Abcam) overnight, followed by incubation with secondary antibody (ab205719; Abcam) for 2 h. The protein bands were visualized with the enhanced chemiluminescence reagent (Vazyme, Nanjing, China).
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from serums, cells and exosomes using TRIzol reagent (Beyotime) and reversely transcribed into cDNAs using PrimeScript™ RT reagent Kit (Takara, Dalian, China) or miRNA 1st Strand cDNA Synthesis Kit (Vazyme). Then, qRT-PCR was conducted with BeyoFast™ SYBR Green qPCR Mix (Beyotime). The expression was calculated with the 2−ΔΔCt method. β-Actin or U6 was utilized as an internal reference. The primers were: circ-ZNF652: 5ʹ -GGGCACAAACAGTTCATGTG-3ʹ and R: 5ʹ-TGCGTTTGAATGATTTTCCA-3ʹ); ZNF652: 5ʹ -CTTCACCAGCAAACAGACTGTGAA-3ʹ and R: 5ʹ-TTCTTTTCTGCATATCCATGGACG-3ʹ); miR-29a-3p: (F: 5ʹ-AGCACCAUCUGAAAUCGGUUA-3ʹ and R: 5ʹ-GTGCAGGGTCCGAGGT-3ʹ); GUCD1: (F: 5ʹ-AGGAGCATCTGGACCATC-3ʹ and R: 5ʹ-GTTCTTGTAGCCCTTGTCG-3ʹ); β-actin: (F: 5ʹ-TGGACTTCGAGCAGGAAATGG-3ʹ and R: 5ʹ-ACGTCGCACTTCATGATCGAG-3ʹ); U6: (F: 5ʹ-CTCGCTTCGGCAGCACA-3ʹ and R: 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ).
RNase R Digestion
Total RNA was added with RNase R (Solarbio) and kept for 15 min at 37°C. Total RNA that was not treated with RNase R (Solarbio) was used as control (Mock). Then, qRT-PCR assay was employed to examine the expression of ZNF652 mRNA and circ-ZNF652.
Subcellular Fraction
The Cytoplasmic & Nuclear RNA Purification Kit (Norgen Biotek, Thorold, Canada) was adopted to separate the nuclear and cytoplasmic RNA in HCC cells according to the protocols. β-Actin was served as a control of cytoplasm transcript and U6 was served as a control of nuclear transcript. Circ-ZNF652 expression was determined via qRT-PCR.
Cell Transfection
Small interfering RNA targeting circ-ZNF652 (si-circ-ZNF652) and its control (si-NC), miR-29a-3p mimic and miRNA NC, miR-29a-3p inhibitor and inhibitor NC, GUCD1 overexpression vector (pc-GUCD1) and pc-Control, lentivirus-mediated short hairpin RNA against circ-ZNF652 (sh-circ-ZNF652) and its control (sh-NC) were synthesized by GeneCopoeia (Guangzhou, China). Then, cell transfection was performed using Lipofectamine 2000 regent (Invitrogen, Carlsbad, CA, USA).
3-(4, 5-Dimethyl-2-Thiazolyl)-2, 5-Diphenyl-2-H-Tetrazolium Bromide (MTT) Assay
The proliferation of SNU-387 and Huh7 cells was assessed using MTT assay. Cells were seeded into 96-well plates and incubated for 0 h, 24 h, 48 h and 72 h. At indicated times, 20 μL MTT (5 mg/mL; Sangon, Shanghai, China) was added into each well and kept for an additional 4 h. Then, the formazan crystals were dissolved with dimethyl sulfoxide (DMSO; Solarbio). The optical density at 490 nm was examined with a microplate reader (BioTek, Winooski, VT, USA).
Transwell Assay
The migration and invasion of HCC were determined using a transwell insert (Corning Incorporated, Corning, NY, USA) pre-coated with (invasion) or without (migration) Matrigel (Solarbio). Briefly, SNU-387 and Huh7 cells were suspended in RPMI 1640 medium (Gibco) and seeded into the upper chamber. RPMI 1640 medium (Gibco) including 10% FBS (Gibco) was added into the bottom chamber. After 24 h, migrated and invaded cells were fixed with methanol and then stained with crystal violet (Solarbio). At last, the stained cells were counted under a light microscope (Olympus, Tokyo, Japan).
Glucose Consumption, Pyruvate Accumulation, Lactate Production and ATP Generation Assays
The levels of glucose consumption, pyruvate accumulation, lactate production and ATP generation were determined using Glucose Assay Kit (Sigma-Aldrich, St. Louis, MO, USA), Pyruvate Assay Kit (Sigma-Aldrich), Lactate Assay Kit (Sigma-Aldrich) and ATP Assay Kit (Sigma-Aldrich), respectively. The experiments were conducted based on the manufacturer’s instructions.
Dual-Luciferase Reporter Assay
The fragments of circ-ZNF652 and GUCD1 3ʹUTR containing the predicted binding sites of wild-type or mutant miR-29a-3p were cloned into the pmirGLO vector (Promega, Fitchburg, WI, USA) to establish the luciferase reporter vectors WT-circ-ZNF652, MUT-circ-ZNF652, WT-GUCD1-3ʹUTR and MUT-GUCD1-3ʹUTR, respectively. Then, the vector was transfected into 293T cells in combination with miRNA NC or miR-29a-3p mimic. At last, the luciferase activity was examined through Dual-Luciferase Reporter Assay Kit (Promega).
RNA Immunoprecipitation (RIP) Assay
RIP assay was conducted to explore the association between circ-ZNF652 and miR-29a-3p using the Magna RIPTM RNA Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA, USA). In brief, SNU-387 and Huh7 cells were lysed in RIP buffer and incubated with magnetic beads which were conjugated with antibody against Argonaute2 (Anti-Ago2; Abcam) or immunoglobulin G (anti-IgG; Abcam). The enrichment of circ-ZNF652 and miR-29a-3p in the beads was determined using qRT-PCR.
Murine Xenograft Model
The animal experiment was performed in accordance with the Guidelines for Care and Use of Laboratory Animals of “National Institutes of Health” and approved by the Ethics Committee of Animal Research of China–Japan Union Hospital of Jilin University. Sh-NC or sh-circ-ZNF652-transfected SNU-387 cells were injected into the nude mice (4–6 weeks old; Shanghai SLAC Laboratory Animals Co., Ltd, Shanghai, China). Seven days later, tumor volume was monitored every 4 days and calculated using the formula: (length×width2)/2. After 27 days, the mice were sacrificed, and tumors were weighed and saved at −80°C for further experiments.
Statistical Analysis
The experiments were repeated three times. The collected data were analyzed by GraphPad Prism 7 software (GraphPad Inc., La Jolla, CA, USA) and displayed as mean ± standard deviation (SD). Student’s t-test or one-way analysis of variance (ANOVA) was used for different analysis. P-value less than 0.05 was considered significantly different.
Results
Exosomal Circ-ZNF652 Was Upregulated in HCC Patients’ Serums and HCC Cells
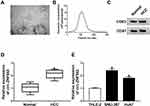
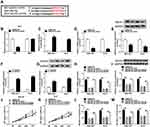
In order to analyze the effect of circ-ZNF652 packaged in exosomes, we first isolated exosomes from HCC patients’ serums and HCC cells. Then, the morphology and size distribution of these isolated particles were identified by TEM and NTA assays. As observed by TEM, the particles showed typical cup-shaped vesicles (Figure 1A). NTA assay showed that the diameters of the most particles were 80 nm-140 nm (Figure 1B). Moreover, exosomal markers (CD63 and CD81) could be detected in the particles (Figure 1C). All these data suggested that the isolated particles were identified as exosomes. Subsequently, we determined the expression of circ-ZNF652 in the exosomes derived from the serums of HCC patients and healthy control by qRT-PCR. The data exhibited that circ-ZNF652 was conspicuously increased in the exosomes from HCC patients’ serums compared to normal controls (Figure 1D). Likewise, the expression of circ-ZNF652 in the exosomes derived from SNU-387 and Huh7 cells was higher than that in the exosomes derived from THEL-2 cells (Figure 1E). To sum up, exosomal circ-ZNF652 might be involved in the progression of HCC.
Silencing of Exosome-Mediated Circ-ZNF652 Suppressed Cell Proliferation, Migration and Invasion in HCC Cells
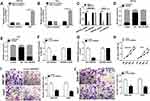
To investigate whether circ-ZNF652 packaged in exosomes could transfer into HCC cells to exert its functions, SNU-387 and Huh7 cells were co-incubated with exosomes isolated from serums, and then circ-ZNF652 expression in SNU-387 and Huh7 cells was measured. We found that circ-ZNF652 was markedly elevated in SNU-387 and Huh7 cells after co-incubation, while the level of circ-ZNF652 in the culture medium was not changed (Figure 2A and B). The results indicated that circ-ZNF652 secreted by exosomes could deliver to HCC cells. Next, we examined the stability of circ-ZNF652 by RNase R digestion assay and the data displayed that RNase R treatment distinctly suppressed ZNF652 mRNA level in SNU-387 and Huh7 cells, but the level of circ-ZNF652 was not affected, indicating that circ-ZNF652 was more stable than liner mRNA of ZNF652 (Figure 2C). Subcellular fraction assay manifested that circ-ZNF652 was mainly located in the cytoplasm in SNU-387 and Huh7 cells (Figure 2D and E). Subsequently, si-circ-ZNF652 was transfected into SNU-387 and Huh7 cells to knock down circ-ZNF652 expression. As exhibited in Figure 2F and G, circ-ZNF652 was effectively decreased in SNU-387 and Huh7 cells following si-circ-ZNF652 transfection, while ZNF652 mRNA level was not changed. The data of MTT assay suggested that circ-ZNF652 knockdown obviously repressed the proliferation of SNU-387 and Huh7 cells compared to si-NC group (Figure 2H). Moreover, the migration and invasion of SNU-387 and Huh7 cells were drastically inhibited by circ-ZNF652 silencing in reference to si-NC group, as illustrated by transwell assay (Figure 2I and J). All these data demonstrated that exosomal circ-ZNF652 could transfer to HCC cells to regulate cell progression.
Circ-ZNF652 Knockdown Suppressed Glycolysis in HCC Cells
It has been reported that hyperglycolysis is a prominent feature of energy metabolism in tumor cells.19 Hence, we explored the effect of circ-ZNF652 on glycolysis in HCC cells by detecting glucose consumption, pyruvate level, lactate production, ATP production and HK2 expression. The data indicated that circ-ZNF652 deficiency drastically reduced glucose consumption (Figure 3A), pyruvate level (Figure 3B), lactate production (Figure 3C) and ATP generation (Figure 3D) in SNU-387 and Huh7 cells in reference to si-NC group, as determined by indicated kit. Moreover, the level of glycolytic related protein HK2 was measured by Western blot assay. The data showed that circ-ZNF652 knockdown led to a remarkable decrease in HK2 expression in SNU-387 and Huh7 cells (Figure 3E). Collectively, circ-ZNF652 knockdown suppressed the level of glycolysis in HCC cells.
Circ-ZNF652 Knockdown Suppressed HCC Cell Proliferation, Migration and Invasion by Targeting miR-29a-3p
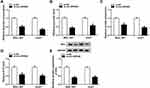
To determine the underlying mechanism of circ-ZNF652 in the regulation of HCC, online software starBase was utilized to search the potential target of circ-ZNF652. As displayed in Figure 4A, circ-ZNF652 contained the potential binding sites of miR-29a-5p. Then, dual-luciferase reporter assay and RIP assay were adopted to verify the targeting association between circ-ZNF652 and miR-29a-3p. Dual-luciferase reporter assay presented that co-transfection of WT-circ-ZNF652 and miR-29a-3p mimic led to a significant inhibition of the luciferase activity in 293T cells compared to WT-circ-ZNF652 and miRNA NC co-transfected group, while no change was observed in MUT-circ-ZNF652 groups (Figure 4B). RIP assay exhibited that circ-ZNF652 and miR-29a-3p were enriched in Anti-Ago2 immunoprecipitates in SNU-387 and Huh7 cells compared to anti-IgG control groups (Figure 4C and D). Moreover, miR-29a-3p was notably increased in SNU-387 and Huh7 cells following circ-ZNF652 deficiency relative to control groups (Figure 4E). These data indicated that circ-ZNF652 knockdown could promote miR-29a-3p expression via targeting miR-29a-3p in HCC cells.
Subsequently, we transfected miR-29a-3p inhibitor or inhibitor NC into SNU-387 and Huh7 cells and found miR-29a-3p expression in SNU-387 and Huh7 cells was dropped after miR-29a-3p inhibitor transfection (Figure 4F). MTT assay demonstrated that the inhibitory effect of circ-ZNF652 silencing on the proliferation of SNU-387 and Huh7 cells was restored by miR-29a-3p inhibitor (Figure 4G and H). As suggested by transwell assay, cell migration and invasion were dramatically repressed by circ-ZNF652 silencing in SNU-387 and Huh7 cells, while the impacts were weakened following miR-29a-3p inhibitor transfection compared to inhibitor NC transfection (Figure 4I and J). Together, circ-ZNF652 knockdown suppressed cell proliferation, migration and invasion via enhancing miR-29a-3p expression in HCC cells.
MiR-29a-3p Depletion Ameliorated the Inhibitory Effect of Circ-ZNF652 Knockdown on Glycolysis in HCC Cells
To explore the relationship between miR-29a-3p and circ-ZNF652 in the regulation of glycolysis in HCC cells, SNU-387 and Huh7 cells were transfected with si-NC, si-circ-ZNF652, si-circ-ZNF652+inhibitor NC or si-circ-ZNF652+miR-29a-3p inhibitor. As determined by specific kits, glucose uptake (Figure 5A), pyruvate level (Figure 5B), lactate production (Figure 5C) and ATP production (Figure 5D) in SNU-387 and Huh7 cells were all inhibited by the deficiency of circ-ZNF652, whereas miR-29a-3p inhibition partly reversed the impacts. Western blot assay showed that circ-ZNF652 silencing caused a marked decrease in HK2 expression in SNU-387 and Huh7 cells, but inhibition of miR-29a-3p overturned the decrease (Figure 5E). Taken together, the suppressive role of circ-ZNF652 knockdown in glycolysis was rescued by miR-29a-3p inhibition in HCC cells.
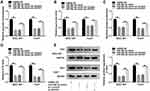
MiR-29a-3p Suppressed Cell Proliferation, Migration and Invasion by Targeting GUCD1 in HCC Cells
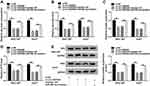
Through further searching starBase, we found that GUCD1 might be a target gene of miR-29a-3p and their potential binding sequences are presented in Figure 6A. Dual-luciferase reporter assay implicated that the luciferase activity was suppressed in 293T cells after miR-29a-3p mimic and WT-GUCD1-3ʹUTR co-transfection in reference to that in cells transfected with miRNA NC and WT-GUCD1-3ʹUTR; however, the luciferase activity was not changed in MUT-GUCD1-3ʹUTR groups (Figure 6B). Then, we transfected miRNA NC or miR-29a-3p mimic into SNU-387 and Huh7 cells. We found that miR-29a-3p was upregulated (Figure 6C), while GUCD1 mRNA and GUCD1 protein were downregulated (Figure 6D and E) in SNU-387 and Huh7 cells transfected with miR-29a-3p mimic compared to those in miRNA NC-transfected cells. Moreover, pc-GUCD1 transfection resulted in a notable elevation in the mRNA and protein levels of GUCD1 in SNU-387 and Huh7 cells in reference to pc-Control group (Figure 6F and G). To explore the association between miR-29a-3p and GUCD1 in the progression of HCC, SNU-387 and Huh7 cells were assigned to miRNA NC, miR-29a-3p mimic, miR-29a-3p mimic+pc-Control and miR-29a-3p+pc-GUCD1 groups. As exhibited in Figure 6H and I, miR-29a-3p elevation markedly reduced the mRNA and protein levels of GUCD1 in SNU-387 and Huh7 cells, while GUCD1 overexpression effectively abrogated the reduction. As revealed by MTT assay, the proliferation of SNU-387 and Huh7 cells was hampered by miR-29a-3p, while GUCD1 overexpression partially abolished the effect (Figure 6J and K). Transwell assay revealed that miR-29a-3p caused a remarkable inhibition in the migration and invasion of SNU-387 and Huh7 cells, but the upregulation of GUCD1 rescued the influences (Figure 6L and M). All these results suggested that GUCD1 reversed the effect of miR-29a-3p on HCC cell progression via acting a target gene of miR-29a-3p.
GUCD1 Overexpression Weakened the Suppressive Role of miR-29a-3p in Glycolysis in HCC Cells
To explore whether miR-29a-3p could modulate glycolysis by targeting GUCD1, SNU-387 and Huh7 cells were divided into four groups: miRNA NC, miR-29a-3p mimic, miR-29a-3p mimic+pc-Control and miR-29a-3p+pc-GUCD1. Next, the level of glycolysis was evaluated by the detection of glucose consumption, pyruvate level, lactate production, ATP level and HK2 expression in SNU-387 and Huh7 cells. As shown in Figure 7A-E, glucose consumption, pyruvate level, lactate production, ATP level and HK2 expression in SNU-387 and Huh7 cells were all decreased by miR-29a-3p, whereas GUCD1 overexpression reversed the effects. These data indicated that miR-29a-3p suppressed glycolysis level in HCC cells, while GUCD1 abolished the suppression.
Circ-ZNF652 Knockdown Suppressed GUCD1 Expression via Sponging miR-29a-3p in HCC Cells
Afterward, we investigated the relationship among circ-ZNF652, miR-29a-3p and GUCD1. As exhibited in Figure 8A and B, knockdown of circ-ZNF652 decreased the mRNA and protein levels of GUCD1 in SNU-387 and Huh7 cells, while GUCD1 overexpression partly restored the decrease. Furthermore, the reduction of GUCD1 mRNA and protein expression in SNU-387 and Huh7 cells caused by circ-ZNF652 silencing was abolished by miR-29a-3p inhibitor (Figure 8C and D). All these data demonstrated that circ-ZNF652 negatively modulated GUCD1 expression by sponging miR-29a-3p in HCC cells.
Circ-ZNF652 Knockdown Hampered Tumor Growth in vivo
To investigate the effect of circ-ZNF652 in vivo, a xenograft tumor mouse model was constructed by injecting sh-NC or sh-circ-ZNF652-transfected SNU-387 cells into the nude mice. As presented in Figure 9A and B, tumor volume and weight were drastically blocked by sh-circ-ZNF652 compared to sh-NC. Moreover, the expression levels of circ-ZNF652, miR-29a-3p and GUCD1 in the tumors were measured. The data indicated that circ-ZNF652 (Figure 9C), GUCD1 mRNA (Figure 9E) and GUCD1 protein (Figure 9F) levels were all reduced, and miR-29a-3p level (Figure 9D) was elevated in the tumor samples collected from the mice in sh-circ-ZNF652 group compared to sh-NC group. In a word, silencing of circ-ZNF652 inhibited tumorigenesis in vivo.
Discussion
In the recent, circRNAs contained in exosomes have been demonstrated to be potential biomarkers of human cancers and play vital roles in tumor progression. For example, exosomal circ_0044516 contributed to prostate cancer cell growth and metastasis.20 Exosomal circRNA_100284 facilitated cell cycle process and cell growth, and caused malignant transformation in human hepatic cells.21 Herein, we mainly determined the effect of exosomal circ-ZNF652 on HCC cell progression. We found exosomal circ-ZNF652 was elevated in HCC patients and HCC cells. Further investigation revealed that circ-ZNF652 silencing repressed HCC cell proliferation, migration, invasion and glycolysis via modulating miR-29a-3p/GUCD1 axis.
It has been announced that diverse circRNAs serve as important regulators in HCC. For instance, circMTO1 was reduced in HCC and its silencing enhanced HCC cell growth and invasion in vitro and promoted tumorigenesis in vivo.22 Circ_0067934 was elevated in HCC and its depletion could decelerate HCC development via inhibiting HCC cell metastasis and growth.23 Moreover, circ_ZNF652 was raised in HCC and circ-ZNF652 deficiency markedly repressed HCC cell metastasis and epithelial–mesenchymal transition (EMT).11 In this research, we found exosomal circ-ZNF652 was elevated in HCC patients’ serums and cells. Then, we co-incubated HCC cells with exosomes derived from HCC patients and found that circ-ZNF652 could deliver to HCC cells. Afterward, loss-of-function experiments were conducted to explore the effect of circ-ZNF652. The data displayed that circ-ZNF652 silencing hampered HCC cell growth and metastasis. Emerging evidence has demonstrated that the accelerated glucose metabolism is a prominent feature of energy metabolism in tumor cells and is associated with the progression of many types of tumors.24 In addition, circRNAs exert important function in regulating cellular metabolism.25 Thus, we investigated the impact of circ-ZNF652 on glycolysis in HCC. We observed that circ-ZHF652 knockdown caused an inhibition in glucose uptake, pyruvate level, lactate production, ATP generation and HK2 expression in HCC cells, suggesting that glycolysis level was inhibited by circ-ZNF652 knockdown.
Since circRNAs can act as miRNA sponges to exert their biological functions,26 we explored the target of circ-ZNF652 and verified that miR-29a-3p was a target of circ-ZNF652. Kong et al disclosed that circFOXO3 acted as a sponge of miR-29a-3p to facilitate prostate cancer cell proliferation and repressed apoptosis.27 In this paper, the influences of circ-ZNF652 silencing on HCC cell growth, metastasis and glycolysis were all weakened by the inhibition of miR-29a-3p, indicating that circ-ZNF652 could sponge miR-29a-3p to modulate HCC progression. In HCC, miR-29a-3p could hamper cell growth and migration by targeting IGF1R.28 Ma et al manifested that miR-29a-3p inhibited HCC cell growth by interacting with PTEN.29 Here, GUCD1 was confirmed to be a target gene of miR-29a-3p. He et al unraveled that GUCD1 restored the suppressive effects of miR-29a-3p on cell metastasis and EMT in HCC.30 Consistently, we found that GUCD1 overexpression abrogated the impacts of miR-29a-3p on cell growth, metastasis and glycolysis. The data illustrated that miR-29a-3p could decelerate HCC cell development via targeting GUCD1.
In summary, the paper illuminated that circ-ZNF652 was elevated in the exosomes derived from HCC patients and HCC cells. Exosome-mediated circ-ZNF652 contributed to cell growth, metastasis and glycolysis via miR-29a-3p/GUCD1 axis. Our findings facilitated our understanding of HCC progression and might support a novel molecular therapeutic target for HCC.
Highlights
- Exosomal circ-ZNF652 is increased in HCC patients’ serums and HCC cells.
- Circ-ZNF652 knockdown suppresses HCC cell proliferation, migration, invasion and glycolysis.
- MiR-29a-3p inhibition reverses the inhibitory effect of circ-ZNF652 silencing on HCC cell progression.
- The suppressive role of miR-29a-3p in HCC cell progression is restored by GUCD1 overexpression.
- Circ-ZNF652 silencing decreases GUCD1 expression via sponging miR-29a-3p.
- Circ-ZNF652 knockdown hinders tumor growth in vivo.
Disclosure
The authors report no funding and no conflicts of interest in this work.
References
1. Bisceglie AM, Rustgi VK, Hoofnagle JH. Hepatocellular carcinoma. Nat Rev Dis Primers. 2016;2:16019.
2. Cidon EU. Systemic treatment of hepatocellular carcinoma: past, present and future. World J Hepatol. 2017;9(18):797–807. doi:10.4254/wjh.v9.i18.797
3. Schwabe RF, Wang TC. Targeting liver cancer: first steps toward a miRacle? Cancer Cell. 2011;20(6):698–699. doi:10.1016/j.ccr.2011.11.021
4. Azmi AS, Bao B, Sarkar FH. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev. 2013;32(3–4):623–642. doi:10.1007/s10555-013-9441-9
5. Xu R, Rai A, Chen M, et al. Extracellular vesicles in cancer - implications for future improvements in cancer care. Nat Rev Clin Oncol. 2018;15(10):617–638. doi:10.1038/s41571-018-0036-9
6. Kahlert C, Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J Mol Med (Berl). 2013;91(4):431–437. doi:10.1007/s00109-013-1020-6
7. Shang Q, Yang Z, Jia R, et al. The novel roles of circRNAs in human cancer. Mol Cancer. 2019;18(1):6. doi:10.1186/s12943-018-0934-6
8. Wang W, Li Y, Li X, et al. Circular RNA circ-FOXP1 induced by SOX9 promotes hepatocellular carcinoma progression via sponging miR-875-3p and miR-421. Biomed Pharmacother. 2019;121:109517. doi:10.1016/j.biopha.2019.109517
9. Liu L, Qi X, Gui Y, et al. Overexpression of circ_0021093 circular RNA forecasts an unfavorable prognosis and facilitates cell progression by targeting the miR-766-3p/MTA3 pathway in hepatocellular carcinoma. Gene. 2019;714:143992. doi:10.1016/j.gene.2019.143992
10. Xu L, Feng X, Hao X, et al. CircSETD3 (Hsa_circ_0000567) acts as a sponge for microRNA-421 inhibiting hepatocellular carcinoma growth. J Exp Clin Cancer Res. 2019;38(1):98. doi:10.1186/s13046-019-1041-2
11. Guo J, Duan H, Li Y, et al. A novel circular RNA circ-ZNF652 promotes hepatocellular carcinoma metastasis through inducing snail-mediated epithelial-mesenchymal transition by sponging miR-203/miR-502-5p. Biochem Biophys Res Commun. 2019;513(4):812–819. doi:10.1016/j.bbrc.2019.03.214
12. Wang Y, Liu J, Ma J, et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer. 2019;18(1):116. doi:10.1186/s12943-019-1041-z
13. Nana-Sinkam SP, Croce CM. MicroRNA regulation of tumorigenesis, cancer progression and interpatient heterogeneity: towards clinical use. Genome Biol. 2014;15(9):445. doi:10.1186/s13059-014-0445-8
14. Reddy KB. MicroRNA (miRNA) in cancer. Cancer Cell Int. 2015;15(1):38. doi:10.1186/s12935-015-0185-1
15. Cheng Y, Qiu L, He GL, et al. MicroRNA-361-5p suppresses the tumorigenesis of hepatocellular carcinoma through targeting WT1 and suppressing WNT/beta-cadherin pathway. Eur Rev Med Pharmacol Sci. 2019;23(20):8823–8832. doi:10.26355/eurrev_201910_19277
16. Gao W, Lu YX, Wang F, et al. miRNA-217 inhibits proliferation of hepatocellular carcinoma cells by regulating KLF5. Eur Rev Med Pharmacol Sci. 2019;23(18):7874–7883. doi:10.26355/eurrev_201909_18997
17. Xiao Z, Wang Y, Ding H. XPD suppresses cell proliferation and migration via miR-29a-3p-Mdm2/PDGF-B axis in HCC. Cell Biosci. 2019;9(1):6. doi:10.1186/s13578-018-0269-4
18. Bellet MM, Piobbico D, Bartoli D, et al. NEDD4 controls the expression of GUCD1, a protein upregulated in proliferating liver cells. Cell Cycle. 2014;13(12):1902–1911. doi:10.4161/cc.28760
19. Munkley J, Elliott DJ. Hallmarks of glycosylation in cancer. Oncotarget. 2016;7(23):35478. doi:10.18632/oncotarget.8155
20. Li T, Sun X, Chen L. Exosome circ_0044516 promotes prostate cancer cell proliferation and metastasis as a potential biomarker. J Cell Biochem. 2019;2.
21. Dai X, Chen C, Yang Q, et al. Exosomal circRNA_100284 from arsenite-transformed cells, via microRNA-217 regulation of EZH2, is involved in the malignant transformation of human hepatic cells by accelerating the cell cycle and promoting cell proliferation. Cell Death Dis. 2018;9(5):454. doi:10.1038/s41419-018-0485-1
22. Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151–1164. doi:10.1002/hep.29270
23. Zhu Q, Lu G, Luo Z, et al. CircRNA circ_0067934 promotes tumor growth and metastasis in hepatocellular carcinoma through regulation of miR-1324/FZD5/Wnt/beta-catenin axis. Biochem Biophys Res Commun. 2018;497(2):626–632. doi:10.1016/j.bbrc.2018.02.119
24. Chen EI. Mitochondrial dysfunction and cancer metastasis. J Bioenerg Biomembr. 2012;44(6):619–622. doi:10.1007/s10863-012-9465-9
25. Yu T, Wang Y, Fan Y, et al. CircRNAs in cancer metabolism: a review. J Hematol Oncol. 2019;12(1):90. doi:10.1186/s13045-019-0776-8
26. Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17(1):79. doi:10.1186/s12943-018-0827-8
27. Kong Z, Wan X, Lu Y, et al. Circular RNA circFOXO3 promotes prostate cancer progression through sponging miR-29a-3p. J Cell Mol Med. 2019;3:2154.
28. Wang X, Liu S, Cao L, et al. miR-29a-3p suppresses cell proliferation and migration by downregulating IGF1R in hepatocellular carcinoma. Oncotarget. 2017;8(49):86592–86603. doi:10.18632/oncotarget.21246
29. Ma JH, Bu X, and Wang JJ, et al. MicroRNA-29-3p regulates hepatocellular carcinoma progression through NF-kappaB pathway. Clin Lab. 2019;65(5). doi:10.7754/Clin.Lab.2018.181012.
30. He Y, He X. MicroRNA-370 regulates cell epithelial-mesenchymal transition, migration, invasion, and prognosis of hepatocellular carcinoma by targeting GUCD1. Yonsei Med J. 2019;60(3):267–276. doi:10.3349/ymj.2019.60.3.267
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.