Back to Journals » Therapeutics and Clinical Risk Management » Volume 14
Examining hemodialyzer membrane performance using proteomic technologies
Authors Bonomini M , Pieroni L , Di Liberato L, Sirolli V, Urbani A
Received 5 September 2017
Accepted for publication 30 October 2017
Published 18 December 2017 Volume 2018:14 Pages 1—9
DOI https://doi.org/10.2147/TCRM.S150824
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Garry Walsh
Mario Bonomini,1 Luisa Pieroni,2 Lorenzo Di Liberato,1 Vittorio Sirolli,1 Andrea Urbani2,3
1Department of Medicine, G. d’Annunzio University, Chieti, 2Proteomic and Metabonomic Units, IRCCS S. Lucia Foundation, Rome, 3Faculty of Medicine, Biochemistry and Clinical Biochemistry Institute, Catholic University of the “Sacred Heart”, Rome, Italy
Abstract: The success and the quality of hemodialysis therapy are mainly related to both clearance and biocompatibility properties of the artificial membrane packed in the hemodialyzer. Performance of a membrane is strongly influenced by its interaction with the plasma protein repertoire during the extracorporeal procedure. Recognition that a number of medium–high molecular weight solutes, including proteins and protein-bound molecules, are potentially toxic has prompted the development of more permeable membranes. Such membrane engineering, however, may cause loss of vital proteins, with membrane removal being nonspecific. In addition, plasma proteins can be adsorbed onto the membrane surface upon blood contact during dialysis. Adsorption can contribute to the removal of toxic compounds and governs the biocompatibility of a membrane, since surface-adsorbed proteins may trigger a variety of biologic blood pathways with pathophysiologic consequences. Over the last years, use of proteomic approaches has allowed polypeptide spectrum involved in the process of hemodialysis, a key issue previously hampered by lack of suitable technology, to be assessed in an unbiased manner and in its full complexity. Proteomics has been successfully applied to identify and quantify proteins in complex mixtures such as dialysis outflow fluid and fluid desorbed from dialysis membrane containing adsorbed proteins. The identified proteins can also be characterized by their involvement in metabolic and signaling pathways, molecular networks, and biologic processes through application of bioinformatics tools. Proteomics may thus provide an actual functional definition as to the effect of a membrane material on plasma proteins during hemodialysis. Here, we review the results of proteomic studies on the performance of hemodialysis membranes, as evaluated in terms of solute removal efficiency and blood–membrane interactions. The evidence collected indicates that the information provided by proteomic investigations yields improved molecular and functional knowledge and may lead to the development of more efficient membranes for the potential benefit of the patient.
Keywords: mass spectrometry, hemodialysis, end-stage renal disease, protein adsorption, biocompatibility, uremic toxin
Introduction
End-stage renal disease (ESRD) requiring renal replacement therapy is a worldwide public health problem, with the number of patients growing at a rate of ~6%–7% annually. ESRD is characterized by the accumulation of various solutes which are normally excreted by the kidneys, causing adverse biochemical/biologic effects and named uremic toxins.1 Hemodialysis (HD) and related extracorporeal techniques such as hemodiafiltration (HDF), hemofiltration (HF), and HF with endogenous reinfusion (HFR) are by far the most commonly used modes of treating ESRD, targeting patient survival and quality of life. Thanks to application of clinical dialysis, for the first time in the age of modern medicine, the death of a vital organ such as the kidney does not equate to the individual’s death. HD is now safer than it was initially. Despite considerable technical and scientific improvements, however, mortality and morbidity are still high among treated patients.2
The main determinant of results in extracorporeal blood purification therapy is the artificial membrane contained in the hemodialyzer, through its clearance and biocompatibility properties. During the extracorporeal procedure, membranes remove excess water and retain the solutes from the uremic blood. However, it should be borne in mind that removal by the HD membrane is nonspecific. So, while the intended removal of toxic solutes is beneficial to the patient, undesired loss of vital plasma components may also occur, depending on the physicochemical characteristics of the membrane.
Proteins are a prime example of the depuration/depletion dichotomy during extracorporeal blood purification (Figure 1). Recognition that medium–high molecular weight (MW) solutes, including several proteins1,3 and protein-bound molecules,4 are potentially toxic has led to the development of more permeable membranes, in order to ameliorate the morbidity and mortality of dialysis patients.5,6 However, along with high removal of middle or large MW solutes, there may be loss of proteins important for the patient’s health, such as albumin, transferrin, or vitamin D-binding protein – a loss which should be restricted if we need to avoid blood deficiency.
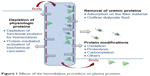  | Figure 1 Effects of the hemodialysis procedure on plasma proteins. |
Removal of blood proteins during HD can occur not only by diffusion and/or convection in the dialysis fluid, but also by adsorption onto the membrane material. Upon exposure to artificial membrane surfaces, adsorption of plasma proteins occurs almost immediately, largely depending on the surface characteristics including roughness, hydrophilicity, chemistry, and charge.7 Some noxious compounds can be removed by high adsorptive membranes.8 Removal by adsorption, however, is again nonselective; moreover, excessive protein adsorption can reduce the filtration capacity and, hence, the therapeutic usefulness of a membrane. Notably, interaction with surface-adsorbed proteins may trigger several biologic pathways including the complement system, the coagulation cascade, and cellular mechanisms.9 So, protein adsorption mainly governs the bio(in)compatibility of a membrane. Bioincompatibility of HD membranes is thought to play a role in both acute and chronic complications associated with extracorporeal blood purification,10 and represents a major challenge to improving the quality of dialysis therapy.11
Notwithstanding the key role played by proteins and protein–protein interactions in all aspects of cell function, until not many years ago, there had been relatively few12 studies in the dialysis setting aiming to identify the complete, or even partial, polypeptide spectrum involved in the process of renal replacement therapy. This deficiency may have been largely related to the lack of reproducible techniques allowing simultaneous assessment of a high number of polypeptides in a single specimen.
Application of proteomic techniques has enabled us to overcome this limitation. Proteomics is being increasingly used and represents a powerful tool affording fundamental and advanced molecular knowledge in the field of uremia and related substitutive therapy,9,13–16 powered in great part by the improvement in proteomic technology. Proteomics can be successfully applied to identification, as well as relative and absolute quantification, of proteins in complex samples such as dialysis fluid or fluid desorbed from the dialysis membrane. In addition, specific software applications enable one to characterize the metabolic pathways and biologic networks affected by the proteins identified, unraveling their pathophysiologic significance. Protein absence/presence and/or concentration may be used to establish a polypeptide pattern for each membrane, which can be compared to that of other membranes. Monitoring the impact on proteins under different dialysis conditions may enhance the quality of renal substitutive therapy and lead to even better care in ESRD patients.9,12
In this article, we review the evidence obtained by proteomic application in the study of hemodialyzer membrane performance, evaluated in terms of solute removal efficiency and blood/membrane interactions (Figure 2). We highlight the most recent and promising findings, showing the current needs and possible future developments in dialysis research.
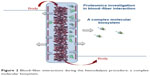  | Figure 2 Blood–fiber interactions during the hemodialysis procedure: a complex molecular biosystem. |
Proteomic analysis of the protein repertoire in dialysis outflow fluid
Early reports on the application of proteomics to the field of dialysis investigated the profile of outflow dialysate obtained from HD using low- or high-flux membranes. Membrane flux categories are based on water permeability, with ultrafiltration coefficients below and above 12 mL/h/mmHg for low- and high-flux membranes, respectively.17
Using capillary electrophoresis coupled to mass spectrometry (MS), Kaiser et al detected 2,515 different polypeptides (based on size and elution time) in the high-flux membrane filtrate and 1,639 in the low-flux membrane filtrate.12 Larger proteins and peptides were present only in the dialysis fluid derived from the high-flux membrane.12 Comparable results were found in a proteomic study by the European Uremic Toxin Working Group.18 Interestingly, these studies indicated a low consensus between the polypeptide profile in dialysis outflow fluid and the urinary proteome profile. This finding suggests that HD cannot substitute all aspects of the kidney function, and that the artificial dialyzer membrane is not comparable with the native kidney.12,18 Similar differences between hemodialysate obtained from low- and high-flux membranes were also demonstrated in a subsequent analysis using both surface-enhanced laser desorption ionization and two-dimensional electrophoresis (2-DE).19 The absence of high-MW proteins from the dialysate obtained with low-flux membranes was confirmed by Western blot analysis.
The broader clearance of proteins overexpressed in uremia19 may lay behind the favorable effects of high-flux membranes on patient outcome, as claimed by clinical studies.20 High-flux membranes are now mostly used in HD units for treatment of ESRD patients because of their better performance. These membranes, however, should not be regarded as a single comparable group, since different removal properties may be detectable even in membranes fabricated from the same biomaterial. This has been shown in a randomized cross-over study comparing three new-generation high-flux polysulfone membranes: Helixone, Amembris, and Purema.21 Sodium dodecyl sulfate-polyacrylamide gel electrophoresis scanning of spent dialysate showed that the three membranes have high efficiency and similar capacities for removal of small-MW compounds, but differ in their ability to remove middle molecules (MW range 8–60 kDa). Significant differences were found between the dialyzers in the amount of proteins removed, particularly in the 20–22.5, 23–30, and 60–80 kDa ranges, with Amembris being associated with the highest protein removal.21 Note that differences in removal among the three membranes identified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis scanning cannot be detected by using other approaches such as single protein determinants and blood measurements, thereby confirming the usefulness of proteomics in evaluating dialysis-associated protein removal.
The lower efficacy of low-flux membranes in the removal of middle-class uremic toxins12,18,19 has been confirmed in a recent randomized cross-over study.22 This study also confirmed21 the presence of significant differences among high-flux membranes in the spectrum of protein removal, with a higher number of protein spots being found by 2-DE in the ultrafiltrate from cellulose triacetate (CTA) in comparison to polyamide and Helixone. Treatment with high-flux membranes in aged patients was associated with excellent safety and clinical tolerability.22 Interestingly, the efficiency of high-flux HD, in particular, with CTA, proved to be similar by means of biochemical and proteomic analyses to on-line HDF. Such observation may be of clinical relevance since on-line HDF, though representing an efficient blood purification treatment, cannot be proposed to all ESRD patients, in particular, the elderly subjects who represent a large proportion of the current dialysis population. Indeed, choice of dialytic treatment in aged patients is quite complex,23 and many of them are treated with low-flux membranes. The study by Donadio et al suggests that aged ESRD patients can be treated safely and efficiently with high-flux membranes, a satisfactory compromise between efficiency and practicability.22
An important effort to identify and characterize uremic toxins, and to steer research toward a better knowledge of the balance between intended and unintended removal of undesired and beneficial proteins, was carried out by Pedrini et al using the multidimensional protein identification technology (MudPIT).24 Two high-flux membranes (Amembris and Polyamix, a polyamide membrane) were used and compared in a randomized cross-over study carried out in 16 patients being dialyzed with high-volume post-dilution HDF. Both membranes displayed high efficiency in removing solutes of different MW in spent dialysate, with Amembris membrane showing higher capability. MudPIT analysis identified 277 proteins in a wide range of MW in the dialysate fluids. Though the protein removal pattern was similar, the total and protein-specific peptide spectral count of most proteins was higher when using Amembris membrane.24 The most abundant proteins identified by MudPIT technology in dialysate included not only the compounds of which the metabolic role and effects of accumulation in the uremic plasma are known,1 such as complement factor D, β2-microglobulin, retinol-binding protein 4, and myoglobin, but also proteins with effects which are less fully described, such as alpha-1 microglobulin/bikunin precursor and prostaglandin D2 synthase. However, vital compounds such as vitamin D-binding protein were also detected in both datasets from the membranes investigated; such depletion may lead to lack of vitamin D, an unwanted condition in HD patients25 that may have a negative impact on survival.26
Proteomics can also be applied for comparatively evaluating performance in terms of removing uremic molecules by newly developed membranes, as compared to those already in use. This is the case with a new polyethersulfone membrane (Synclear 0.2) used for the convective chamber in the HFR dialyzer.27 HFR is an integrated dialysis technique that uses a double chamber dialyzer, a high-flux (convective) filter in series with a low-flux (diffusive) filter, coupled with a sorbent cartridge containing a styrenic resin able to retain uremic toxins from the ultrafiltrate, while amino acids, albumin, and water-soluble vitamins are reinfused into the bloodstream.28 The proteomic profile of the ultrafiltrate obtained during an HFR session in vivo showed that the new membrane had higher purification efficiency for middle–high MW molecules than the conventional ones.27 The proteins identified by electrospray ionization-quadrupole (ESI-Q)-time of flight (TOF) coupled with on-chip elution were mostly uremic retention solutes. Estimation by exponentially modified protein abundance index of the relative abundance of these proteins confirmed better clearance of toxin molecules achieved by Synclear 0.2.27 Thus, unlike standard HD membranes, the new membrane displayed the ability to break the “albumin wall”, efficiently clearing middle–high MW proteins. Loss of important nutrients in the ultrafiltrate may occur, however; so, further studies are needed to optimize the retaining behavior of the cartridge in this promising efficient dialytic approach.
More recently, removal of cell-activating substances from uremic blood was investigated in dialysate samples collected during HD with a high-flux membrane and a medium-cut-off membrane (MCO-CI) which has a higher permeability.29 The latter is a new designed highly porous and selective membrane, characterized by permeability and pore size distribution similar to the kidney itself.30 Treatment of tubular epithelial cells as an experimental model with the MCO-CI dialysate, as compared to high-flux dialysate, was associated with a significant decrease in cell viability and with loss of brick-like shape of the cell and cell–cell connections.29 The observed phenotype is supported by proteomic analysis (liquid chromatography–MS/MS) coupled with bioinformatics tools, of dialysate samples. Functional classification of proteins overrepresented in samples obtained from HD with MCO-CI revealed the presence of many enzymes (oxidoreductases, transferases, isomerases, and enzyme modulators), signaling molecules, cell adhesion molecules, transfer proteins, immunity proteins, and multiple proteins known to affect tubular epithelial cells’ viability and morphology.29 Proteomic findings were linked by toxicity functional analysis to renal, cardiovascular, and liver toxicity. In addition, pathway analysis showed the proteins mostly removed by MCO-CI dialyzer to be involved in inflammatory responses, cellular stress and injury, cellular growth and proliferation, cardiovascular signaling, and nuclear receptor signaling.29 Several of these proteins had been associated with advanced stages of chronic kidney disease,31 which suggests that their removal by MCO-CI dialyzer might improve patient outcome, a concept which requires further studies.
Proteomic analysis of proteins adsorbed onto dialyzer membrane
Protein adsorptive properties are a key feature of membranes used for dialysis therapy.11,32 Protein adsorption influences membrane removal performance and is vital to the biocompatibility of a membrane material. Thus, protein adsorption should be carefully monitored during the development of biomaterials for HD treatment,33 evaluating the composition, amount, and conformational change of the proteins adsorbed.
Though nonselective, adsorption onto the membrane surface can represent an important mechanism for eliminating some toxic compounds such as β2-microglobulin. Circulating levels of β2-microglobulin in HD patients serve as a surrogate marker of middle-molecule uremic toxins, and reduction of them is thought to be beneficial for such patients.34 Clinical studies have indicated the serum β2-microglobulin level as a predictor of mortality in chronic dialysis patients.35 Furthermore, removal of β2-microglobulin may be effective for the treatment of dialysis-associated amyloidosis,34,36 a destructive articular disease, and may reduce the exposure of aminophospholipid phosphatidylserine on the erythrocyte surface,37 an abnormality of pathophysiologic significance in dialysis patients.38,39
Ishikawa et al40 used surface-enhanced laser desorption ionization-TOF MS to investigate proteins and peptides in serum, outflow dialysate, and fluid desorbed from dialysis membrane, during clinical HD with two different membrane materials, polysulfone and polymethylmethacrylate. Both dialyzers were associated with a marked reduction in post-dialysis serum β2-microglobulin concentration. However, while β2-microglobulin was present in a small amount in outflow dialysis fluid and in a large amount in desorbed fluid (indicative of protein adsorption) when using polymethylmethacrylate, the opposite behavior was found with polysulfone. Further specific differences were also found between the two membranes in the profile of proteins filtered and proteins adsorbed.40 This study clearly shows that the relative participation of mechanisms involved in solute removal during HD (filtration, adsorption) may vary according to membrane characteristics. Thus, for a comprehensive and accurate evaluation of the effective solute removal (what has left the plasma) during the extracorporeal procedure, measurements in both the outflow dialysis fluid and the eluate reflecting membrane adsorption are necessary.
Plasma proteins adsorbed onto a membrane surface govern the ensuing biologic reactions which may occur during the extracorporeal procedure and which define the membrane’s bio(in)compatibility. Thus, the quantitative and qualitative evaluation of a material’s affinity for proteins is an essential factor when assessing the performance of the material and the possible development of new biomaterials with improved biocompatibility. Our research group studies over the last decade have shown the suitability of proteomics for investigating protein adsorption in an unbiased manner and in its full complexity.
In our first proof-of-principle investigation, we examined in an in vitro HD system the protein-binding characteristics of two different hollow-fiber minidialyzers composed of cellulose diacetate or ethylene-vinyl alcohol (EVAL).41 The total protein amount in the eluate obtained by the two membrane materials was significantly higher for cellulose diacetate, as was also confirmed in a subsequent study.42 In addition, by 2-DE coupled to nanoLC–MS/MS analysis, several different proteins were identified as common or characteristic of filter material interaction, demonstrating the possibility of identification following isolation of compounds by a proteomic approach.41 Notably, protein adsorptive properties evidenced by proteomics correlated well with certain membrane surface characteristics affecting protein adsorption.7 EVAL, for instance, displayed a more regular lattice (by atomic force microscopy) and higher hydrophilicity (by pulsed nuclear magnetic resonance) than cellulose diacetate. These results suggest the potential support that proteomics may provide for the development of biomaterials.
In a subsequent study, dialyzers used for clinical HD composed of either polysulfone-based Helixone or CTA materials were investigated by an ex vivo apparatus in a parallel set of experiments, the profiling strategy being based on 2-DE separation of desorbed proteins coupled to matrix-assisted laser desorption/ionization-TOF/TOF analysis.43 Protein concentration in the eluate did not differ between the two different membranes. However, the protein compositions, albeit with some similarities, were altogether qualitatively different. The map of retained proteins onto CTA was made up of 239 protein spots, mainly albumin and apolipoproteins. In contrast, proteome maps from Helixone (179 protein spots) showed an abundance of other proteins, particularly fibrinogen isoforms and low-MW fragments of fibrinogen.43 To further examine the specific role of those membrane biomaterials on the adsorption profile of plasma proteins, a prospective cross-over study was carried out in vivo in six HD patients.44 MS analysis and shotgun analysis by nanoLC–MS of proteins eluted from Helixone or CTA membranes at the end of HD substantially confirmed previous findings,43 demonstrating a predominant adsorption of low abundant plasma proteins onto Helixone material and high abundant plasma proteins onto CTA. Importantly, the difference proved to be related to membrane material properties and not to patient characteristics.44
Identification of protein composition in the eluate can give notable insights into the biologic reactions which may take place during dialysis. In proteome maps from Helixone membrane (but not from CTA), the presence of fibrinogen fragments, in addition to intact fibrinogen, both in vitro43 and in vivo44 may indicate the occurrence of fibrinolytic and procoagulatory activity upon blood exposed to the material. The possible relevance of these findings in vivo is suggested by the significant increase in plasma fibrinogen levels in ESRD patients routinely treated with Helixone dialyzer, while CTA was not associated with increases in fibrinogen, as found in a retrospective analysis.43 Moreover, in a more recent study, application of shotgun proteomics data-independent analysis in eluates obtained after clinical HD revealed that Helixone membrane, unlike CTA, is associated with the retention of proteins due to the coagulation cascade and platelet activation.45 An increase in specific coagulation proteins was validated by targeted MS using selected reaction monitoring on proteotypic transitions of key protein effectors. A slightly higher significant platelet activation profile was also observed after patients had undergone HD treatment with Helixone dialyzer, a phenomenon that does not appear as a generalized process but is restricted to certain pathways such as the collagen pathway.45 This evidence, supported by previous findings,46 might indicate a mechanism associated with the physicochemical properties of Helixone material, in particular, a negatively charged surface.43 The lower effect brought about by CTA material might be related to the predominant adsorption of albumin onto its surface,43,44 which is thought to be beneficial owing to this protein’s relative lack of glycosylation, which prevents platelet adhesion.47 Our results are in keeping with previous clinical observations showing the potentially higher activation of the coagulation cascade for polysulfone-based material (such as Helixone) as compared to CTA material.46,48,49
The tendency to activate the coagulation cascade via blood–membrane contact is one measure of the biocompatibility of an HD membrane and requires anticoagulation during the HD procedure to avoid clotting of the extracorporeal circuit. Optimal anticoagulation, however, remains a controversial issue for standard practice,50 and even sophisticated coagulation tests cannot adequately predict the risk of clotting.51 The collected evidence highlights a modulation of the coagulation system during the HD procedure, which is largely influenced by the biomaterial used. Our results43–45 suggest that proteomic data may provide a molecular basis for interpretation of the efficacy and safety of the intradialytic anticoagulation regimen. Our results also suggest that membrane material should probably be taken into account, together with patient characteristics (age, use of anticoagulants, and atherosclerotic vascular disease at risk of plaque rupture) in the anticoagulant prescription for each HD patient. Membrane flux configuration also should necessitate consideration for intradialytic anticoagulation, according to a very recent study by Han et al.52 The authors investigated by proteomics the protein adsorption profile of two membranes made from the same biomaterial (polyamix membrane), but with a different flux configuration (low flux and high flux, low flux having a smaller pore size). Proteins related to the coagulation cascade proved to be differentially adsorbed onto the surfaces of the two membranes. In particular, the α and β chains of fibrinogen, which activated and formed fibrin, were more adsorbed by the high-flux membrane, which might indicate the need for a higher anticoagulation during high-flux dialysis.52
Application of Ingenuity Pathway Analysis system software to datasets of eluates obtained from Helixone or CTA materials showed the involvement of adsorbed proteins in important molecular processes, such as lipid metabolism, molecular transport, cell growth differentiation and communication, and small molecule biochemistry, entailing differences in the ranking networks between the two membranes.45 Most molecules retained by CTA membrane proved to be secreted in the extracellular region, while the corresponding analysis performed on Helixone membrane adsorbed proteins showed both secreted (extracellular) and cellular proteins. In addition, canonical pathway analysis performed to identify the most relevant metabolic and signaling pathways among those present in the dataset53 showed involvement of proteins mostly adsorbed by CTA in the pathway related to activation of LXR/RXR, the retinoid X nuclear receptors mediating the biologic effects of retinoids, while the proteins mostly retained by Helixone filters proved to be involved in the canonical pathways of vascular endothelial growth factor signaling and integrin signaling.45 These analyses represent a further effort to fathom the pathophysiologic importance of protein involvement in HD process.
In the context of protein adsorption pattern investigations, other authors have examined whether a preferential adsorption onto HD membrane might occur. In a study on the potential molecules involved in blood–dialyzer interaction during HD with a low-flux polysulfone membrane, a comparative analysis of the eluate proteome to matched plasma samples from the respective patients showed 10 protein spots with a relative eluate intensity significantly higher than that in plasma.54 These proteins included ficolin-2, complement C3c fragment, clusterin, and apolipoproteins, suggesting preferential adsorption for these proteins, and hence potential engagement in blood–membrane interactions.54 These results were substantially confirmed in a later study by the same group in a larger HD cohort (n=16), among other things showing a significant decrease in ficolin-2 levels, which proved to be correlated with dialysis-induced leukopenia and generation of complement component C5a.55 Ficolin-2 is a humoral molecule with lectin-like activity and has an important role in innate immunity.56 Thus, the lectin pathway initiated by adsorption of ficolin-2 can be involved in complement activation upon contact of blood with the polysulfone membrane, knowledge of which could help us develop better tolerated biodevices.55
In a more recent study, high-performance liquid chromatography coupled to matrix-assisted laser desorption/ionization-TOF MS was used to analyze and compare the protein patterns in plasma, dialysate, and membrane in the same patient undergoing three different extracorporeal blood purification modalities (HD, HF, HDF).57 Plasma protein change and dialysate peptide concentration exhibited small differences among the three dialysis modalities. However, the relative hemoglobin removal ratio, quantified through the membrane protein deposits, proved to be different (about 1.7 for HD, 1.2 for HDF, and 1.0 for HF), thereby confirming the usefulness of proteomics in evaluating the effects of dialysis.57
Conclusion
The widespread use of dialysis to prolong life for people without kidney function has been undoubtedly an achievement of enormous significance. Despite significant technological improvements, however, the results of extracorporeal blood purification therapy for ESRD patients are still not fully satisfactory. The polymer that constitutes the hemodialyzer membrane (the central component of the dialytic process) gives the membrane its characteristic physiochemical properties, which directly affect the membrane interactions with blood components. Overall, the evidence collected indicates that proteomics may be eminently suitable to evaluate membrane performance. Proteomic approaches allow one to characterize the protein removal pattern of any biomaterial used for HD membranes, highlighting the intentional clearance of potential toxic compounds as well as undesired loss of vital proteins. Optimal performing membranes should eliminate accumulated proteins with putative toxic potential, while leaving essential macromolecules undepleted. Again, proteomic application may provide a better understanding of the molecular and submolecular substrates involved in blood–membrane interactions causing plasma adsorption and the subsequent reaction cascade during the extracorporeal procedure.9,55 Thus, proteomic application may provide information relevant to the field of renal substitutive therapy. Improved knowledge about the impact of biomaterial characteristics upon interaction with plasma proteins may lead to the development of more efficient and biocompatible polymers for the potential benefit of the ESRD patient.
Author contributions
All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Duranton F, Cohen G, De Smet R, et al; European UremicToxin Work Group. European Uremic Toxin Work Group. Normal and pathologic concentrations of uremic toxins. J Am Soc Nephrol. 2012;23(7):1258–1270. | ||
Himmelfarb J, Ikizler TA. Hemodialysis. N Engl J Med. 2010;363:1833–1845. | ||
Vanholder R, De Smet R, Glorieux G, et al; European Uremic Toxin Work Group (EUTox). Review on uremic toxins: classification, concentration, and interindividual variability. Kidney Int. 2003;63(5):1934–1943. | ||
Vanholder R, Schepers E, Pletinck A, Nagler EV, Glorieux G. The uremic toxicity of indoxyl sulfate and p-cresyl sulfate: a systematic review. J Am Soc Nephrol. 2014;25(9):1897–1907. | ||
Baurmeister U, Vienken J, Ward RA. Should dialysis modalities be designed to remove specific uremic toxins? Semin Dial. 2009;22(4):454–457. | ||
Lekawanvijit S, Krum H. Cardiorenal syndrome: role of protein-bound uremic toxins. J Ren Nutr. 2015;25(2):149–154. | ||
Werner C, Jacobasch H. Surface characterization of polymers for medical devices. Int J Artif Organs. 1999;22(3):160–176. | ||
Vanholder RC, Eloot S, Glorieux GL. Future avenues to decrease uremic toxin concentration. Am J Kidney Dis. 2016;67(4):664–676. | ||
Bonomini M, Sirolli V, Pieroni L, Felaco P, Amoroso L, Urbani A. Proteomic Investigations into hemodialysis therapy. Int J Mol Sci. 2015;16(12):29508–29521. | ||
Pertosa G, Grandaliano G, Gesualdo L, Schena FP. Clinical relevance of cytokine production in hemodialysis. Kidney Int Suppl. 2000;76:S104–S111. | ||
Chanard J, Lavaud S, Randoux C, Rieu P. New insights in dialysis membrane biocompatibility: Relevance of adsorption properties and heparin binding. Nephrol Dial Transplant. 2003;18(2):252–257. | ||
Kaiser T, Hermann A, Kielstein JT, et al. Capillary electrophoresis coupled to mass spectrometry to establish polypeptide patterns in dialysis fluids. J Chromatogr A. 2003;1013(1–2):157–171. | ||
Thongboonkerd V. Proteomics in extracorporeal blood purification and peritoneal dialysis. J Proteomics. 2010;73(3):521–526. | ||
Bonomini M, Sirolli V, Magni F, Urbani A. Proteomics and nephrology. J Nephrol. 2012;25(6):865–871. | ||
Mullen W, Saigusa D, Abe T, Adamski J, Mischak H. Proteomics and metabolomics as tools to unravel novel culprits and mechanisms of uremic toxicity: instrument or hype? Semin Nephrol. 2014;34(2):180–190. | ||
Vanholder R, Boelaert J, Glorieux G, Eloot S. New methods and technologies for measuring uremic toxins and quantifying dialysis adequacy. Semin Dial. 2015;28(2):114–124. | ||
Chelamcharla M, Leypoldt JK, Cheung AK. Dialyzer membranes as determinants of the adequacy of dialysis. Semin Nephrol. 2005;25(2):81–89. | ||
Weissinger EM, Kaiser T, Meert N, et al. Proteomics: a novel tool to unravel the pathophysiology of uraemia. Nephrol Dial Transplant. 2004;19(12):3068–3077. | ||
Dihazi H, Muller CA, Mattes H, Muller GA. Proteomic analysis to improve adequacy of hemo- and peritoneal dialysis: Removal of small and high molecular weight proteins with high- and low-flux filters or a peritoneal membrane. Proteomics Clin Appl. 2008;2(7–8):1167–1182. | ||
Woods HF, Nandakumar M. Improved outcome for haemodialysis patients treated with high-flux membranes. Nephrol Dial Transplant. 2000;15(Suppl 1):36–42. | ||
Ficheux A, Gayrard N, Szware I, et al. The use of SDS-PAGE scanning of spent dialysate to assess uraemic toxin removal by dialysis. Nephrol Dial Transplant. 2011;26(7):2281–2289. | ||
Donadio C, Kanaki A, Sami N, Tognotti D. High-flux dialysis: clinical, biochemical, and proteomic comparison with low-flux dialysis and on-line hemodiafiltration. Blood Purif. 2017;44(2):129–139. | ||
Berger JR, Jaikaransingh V, Hedayati SS. End-stage kidney disease in the elderly: approach to dialysis initiation, choosing, modality, and predicting outcomes. Adv Chronic Kidney Dis. 2016;23(1):36–43. | ||
Pedrini LA, Krisp C, Gmerek A, Wolters DA. Patterns of proteins removed with high-flux membranes on high-volume hemodiafiltration detected with a multidimensional LC-MS/MS strategy. Blood Purif. 2014;38(2):115–126. | ||
Speeckaert MM, Glorieux GL, Vanholder R, et al. Vitamin D binding protein and the need for vitamin D in hemodialysis patients. J Ren Nutr. 2008;18(5):400–407. | ||
Lin YP, Yang CY, Liao CC, Yu WC, Chi CW, Lin CH. Plasma protein characteristics of long-term hemodialysis survivors. PLoS One. 2012;7(7):e40232. | ||
Cuoghi A, Caiazzo M, Monari E, et al. New horizon in dialysis depuration: Characterization of a polysulfone membrane able to break the “albumin wall”. J Biomater Appl. 2015;29(10):1363–1371. | ||
Wratten ML, Ghezzi PM. Hemodiafiltration with endogenous reinfusion. Contrib Nephrol. 2007;158:94–102. | ||
Latosinska A, Hulko M, Speidel R, Mischak H, Storr M, Krause B. Removal of cell-activating substances using dialyzers with various permeability profiles. Artif Organs. Epub 2017 Jul 26. | ||
Boschetti-de-fierro A, Voigt M, Storr M, Krause B. MCO membranes enhanced selectivity in high-flux class. Sci Rep. 2015;5:18448. | ||
Glorieux G, Mullen W, Duranton F, et al. New insights in molecular mechanisms involved in chronic kidney disease using high-resolution plasma proteome analysis. Nephrol Dial Transplant. 2015;30(11):1842–1852. | ||
Pascual M, Tolkoff-Rubin N, Schifferli JA. Is adsorption an important characteristic of dialysis membranes? Kidney Int. 1996;49(2):309–313. | ||
Vienken J. Polymers in nephrology. Characteristics and needs. Int J Artif Organs. 2002;25(5):470–479. | ||
Lonnemann G, Koch KM. Beta(2)-microglobulin amyloidosis: effects of ultrapure dialysate and type of dialyzer membrane. J Am Soc Nephrol. 2002;13(Suppl 1):S72–S77. | ||
Okuno S, Ishimura E, Kohno K, et al. Serum beta2-microglobulin level is a significant predictor of mortality in maintenance haemodialysis patients. Nephrol Dial Transplant. 2009;24(2):571–577. | ||
Kazama JJ, Maruyama H, Gejyo F. Reduction of circulating beta2-microglobulin level for the treatment of dialysis-related amyloidosis. Nephrol Dial Transplant. 2001;16(Suppl 4):31–35. | ||
Bonomini M, Ballone E, Di Stante S, et al. Removal of uraemic plasma factor(s) using different dialysis modalities reduces phosphatidylserine exposure in red blood cells. Nephrol Dial Transplant. 2004;19(1):68–74. | ||
Bonomini M, Sirolli V, Reale M, Arduini A. Involvement of phosphatidylserine exposure in the recognition and phagocytosis of uremic erythrocytes. Am J Kidney Dis. 2001;37(4):807–814. | ||
Bonomini M, Pandolfi A, Di Pietro N, et al. Adherence of uremic erythrocytes to vascular endothelium decreases endothelial nitric oxide synthase expression. Kidney Int. 2005;67(5):1899–1906. | ||
Ishikawa I, Chikazawa Y, Sato K, et al. Proteomic analysis of serum, outflow dialysate and adsorbed protein onto dialysis membranes (polysulfone and PMMA) during hemodialysis treatment using SELDI-TOF-MS. Am J Nephrol. 2006;26(4):372–380. | ||
Bonomini M, Pavone B, Sirolli V, et al. Proteomics characterization of protein adsorption onto hemodialysis membranes. J Proteom Res. 2006;5(10):2666–2674. | ||
Pavone B, Sirolli V, Bucci S, et al. Adsorption and carbonylation of plasma proteins by dialyser membrane material: in vitro and in vivo proteomics investigations. Blood Transfus. 2010;8(Suppl 3):s113–s119. | ||
Urbani A, Lupisella S, Sirolli V, et al. Proteomic analysis of protein adsorption capacity of different haemodialysis membrane. Mol BioSyst. 2012;8(4):1029–1039. | ||
Urbani A, Sirolli V, Lupisella S, et al. Proteomic investigations on the effect of different membrane materials on blood protein adsorption during haemodialysis. Blood Transfus. 2012;10(Suppl 2):S101–S112. | ||
Pieroni L, Levi Mortera S, Greco V, et al. Biocompatibility assessment of haemodialysis membrane materials by proteomic investigations. Mol Biosyst. 2015;11(6):1633–1643. | ||
Kuragano T, Kuno T, Takahashi Y, et al. Comparison of the effects of cellulose triacetate and polysulfone membrane on GPIIb/IIIa and platelet activation. Blood Purif. 2003;21(2):176–182. | ||
Oleschuk RD, McComb ME, Chow A, et al. Characterization of plasma proteins adsorbed onto biomaterials by MALDI-TOFMS. Biomaterials. 2000;21(16):1701–1710. | ||
Olafiranye F, Kyaw W, Olafiranye O. Resolution of dialyzer membrane-associated thrombocytopenia with use of cellulose triacetate membrane: a case report. Case Rep Med. 2011;2011:134295. | ||
Liu S, Shi W, Liang X, et al. Cellulose triacetate dialyser reduces platelet loss during continuous veno-venous hemofiltration. Blood Purif. 2010;29(4):375–382. | ||
Ouseph R, Ward RA. Anticoagulation for intermittent hemodialysis. Semin Dial. 2000;13(3):181–187. | ||
Sagedal S, Hartmann A, Sundstrom K, Bjørnsen S, Brosstad F. Anticoagulation intensity sufficient for haemodialysis does not prevent activation of coagulation and platelets. Nephrol Dial Transplant. 2001;16(5):987–993. | ||
Han S, Yang K, Sun J, Liu J, Zhang L, Zhao J. Proteomics investigations into serum proteins adsorbed by high-flux and low-flux dialysis membranes. Proteomics Clin Appl. Epub 2017 Aug 10. | ||
Ciavardelli D, D’Orazio M, Pieroni L, et al. Proteomic and ionomic profiling reveals significant alterations of protein expression and calcium homeostasis in cystic fibrosis cells. Mol BioSyst. 2013;9(6):1117–1126. | ||
Mares J, Thongbookerd V, Tumaz Z, Moravec J, Matejovic M. Specific adsorption of some complement activation proteins to polysulfone dialysis membranes during hemodialysis. Kidney Int. 2009;76(4):404–413. | ||
Mares J, Richtrova P, Hricinova A, et al. Proteomic profiling of blood-dialyzer interactome reveals involvement of lectin complement pathway in hemodialysis-induced inflammatory response. Proteomics Clin Appl. 2010;4(10–11):829–838. | ||
Runza VL, Schwaeble W, Mannel DN. Ficolins: novel pattern recognition molecules of the innate immune response. Immunobiology. 2008;213(3–4):297–306. | ||
Naseeb U, Zarina S, Jagerbrink T, Shafqat J, Jörnvall H, Axelsson J. Differential hemoglobin A sequestration between hemodialysis modalities. Biomol Concepts. 2017;8(2):125–129. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
