Back to Journals » International Journal of General Medicine » Volume 14
Evaluation of Analytical Performance of Seven Rapid Antigen Detection Kits for Detection of SARS-CoV-2 Virus
Authors Eshghifar N, Busheri A, Shrestha R , Beqaj S
Received 18 December 2020
Accepted for publication 15 January 2021
Published 12 February 2021 Volume 2021:14 Pages 435—440
DOI https://doi.org/10.2147/IJGM.S297762
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Scott Fraser
Nahal Eshghifar,1 Ali Busheri,1 Rojeet Shrestha,2 Safedin Beqaj1
1UltimateDx Laboratories, Los Angeles, CA, USA; 2Faculty of Health Sciences, Hokkaido University, Sapporo, Japan
Correspondence: Safedin Beqaj
UltimateDx Laboratories, 516 North Larchmont Blvd, Los Angeles, CA, 92620, USA
Email [email protected]
Background: Early diagnosis of the novel coronavirus disease of 2019 (COVID-19) in asymptomatic and symptomatic patients is crucial to identify infectious individuals and to help prevent the spread of the virus in the community. Several assays have been developed and are in use in today’s clinical practice. These assays vary in their analytical and clinical performance. For an accurate diagnosis, medical professionals must become more familiar with the test’s utility to select the most appropriate test. This study aims to evaluate the analytical performance of rapid antigen tests used for the detection of SARS-CoV-2 viral antigen compared to RT-PCR SARS-CoV-2 molecular assay.
Methods: Oropharyngeal swab specimens from five COVID-19 patients were tested by seven rapid antigen tests developed by different IVD companies. RT-PCR to detect specific RNA fragments of SARS-CoV-2 was used as a confirmatory test. The cycle threshold (Ct) value, which often reflects viral load, in these specimens ranged from 15 to 35. For the analytical evaluation, extraction fluid of each antigen kit was spiked with attenuated ATCC virus at different concentrations ranging from 4.6x104/mL to 7.5x105/mL and tested with antigen testing kits.
Results: Out of five confirmed positive SARS-CoV-2 specimens by RT-PCR, only one sample showed a positive result by one of the seven evaluated antigen testing kits. The positive result was observed in the specimen with a Ct value of 15. All other evaluated rapid tests were negative for all five positive specimens. This was further confirmed with the spiking study using ATCC attenuated virus, where extraction fluid of each rapid test was spiked with concentrations ranging from 4.6x104/mL to 7.5x105/mL. None of these spiked specimens showed positive results, indicating very low sensitivity of these antigen kits.
Conclusion: This comparison study shows that rapid antigen tests are less sensitive than RT-PCR tests and are not reliable tests for testing asymptomatic patients, who often carry low viral load. Analytical performance of rapid antigen tests should be thoroughly evaluated before implementing it at clinical decision level.
Keywords: SARS-CoV-2, COVID-19, rapid antigen detection, RT-PCR
Background
Since COVID-19 was declared a global pandemic, the goal of the medical community has been to offer rapid, massive, and affordable testing. To meet this goal, several assays have been developed and are in use in clinical practice. These assays can be broadly categorized based on the testing methodology used for the detection of SARS-CoV-2. These include antibody testing, antigen testing, and molecular testing.1–3 Antibody testing (serology testing) is used for the detection of antibodies to SARS-CoV-2 in blood or serum. It can be total antibodies against the virus or its specific subtypes – IgA, IgG, and IgM. Antigen testing is based on immunoassay, mostly immunoblot technology for the detection of SARS-CoV-2 antigen in the nasopharyngeal or oropharyngeal swab specimens, whereas molecular testing is designed to detect SARS-CoV-2 viral nucleic acid (RNA) using the Nucleic Acid Amplification Test (NAAT).4
There is no doubt that widespread testing for containing the virus and getting the pandemic under control is critical. As the majority of individuals with COVID-19 are asymptomatic or show mild symptoms, laboratory confirmation of SARS-CoV-2 infection is crucial.5 The appropriate test and timing for sampling is also another critical factor for the accurate diagnosis of COVID-19. Numerous tools are readily available for healthcare professionals for guidance on testing. One such tool, Medical Database (MD), a guide developed to assist healthcare professionals, recommends that a molecular test should be used as the gold standard for a diagnostic test to detect the SARS-CoV-2 virus and IgG antibody testing should be used for evaluating the immune response after the infection subsides.6 Accordingly, IgA and IgM antibody testing have no diagnostic value but can be used for the evaluation of severity of disease/infection. Molecular testing for the detection of viral RNA is currently the most widely used diagnostic test. These molecular assays used to diagnose SARS-CoV-2 are designed to amplify and detect specific target viral genes and regions including the spike (S), envelope (E), and nucleocapsid (N) proteins which represent three of the four proteins that structurally constitute the virus as well as the RNA dependent RNA polymerase (RdRp) gene and the Open Reading Frame 1ab (ORF1ab) region.7 Although lower respiratory tract specimens generally have a higher positivity rate, upper respiratory specimens like nasopharyngeal or oropharyngeal swabs are most widely used as the specimen of choice.8
These molecular tests can detect the viral nucleic acid within 1–3 days of exposure or infection, and can be used for early identification of asymptomatic patients.9 Unfortunately, these tests are expensive, and require complex technology and well-trained testing personnel. There is also an issue of turnaround time which may take as long as a few days. To achieve the urgent need for massive testing to control this pandemic, there is a need for an alternative assay which is high-throughput, rapid, simple, and economic. Furthermore, 60% of clinical laboratories around the globe are currently facing supply chain problems with appropriate reagent test kits and consumables.10 To mitigate the drawbacks of molecular testing, viral protein antigen testing of respiratory samples by immunoassay can be a promising alternative. Such antigen-based testing has been recently endorsed by the US Centers for Disease Control and Prevention (CDC) and interim guidance for its use has been formulated by the WHO.11 Many companies have come up with a rapid antigen test as laboratory-based tests and point-of-care tests. To this date (December 7, 2020) only seven of them have been approved by the US FDA under Emergency Use Authorizations (EUA). These antigen-detecting rapid diagnostic tests (Ag-RDT) are based on lateral flow immunochromatographic assay for the detection of nucleocapsid protein antigens specific to SARS-CoV-2. Although such antigen tests offer widespread rapid testing without the need of expensive laboratories, it is very important to understand the analytical sensitivity of these rapid antigen tests. Unfortunately, there is limited information available on the analytical and clinical performance of antigen testing. More studies are required to evaluate the sensitivity and specificity of antigen testing before implementing this assay effectively. In this study, we evaluated the analytical sensitivity of SARS-CoV-2 rapid antigen tests developed by seven IVD companies, of which three are FDA EUA approved, by comparing it to the gold standard molecular assay.
Methods
Specimens
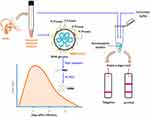
Oropharyngeal swabs were collected from five COVID-19 patients confirmed positive by SARS-CoV-2 RT-PCR assay. The swabs were collected in universal transport media (UTM) (BD Diagnostics, Sparks, MD, USA) and appropriately stored. A schematic diagram of the study methodology is illustrated in Figure 1. Written informed consent was obtained from each patient and the study protocol was approved by the Institutional Ethical Board of UltimateDx Laboratories in agreement with the World Medical Association Declaration of Helsinki.
RT-PCR
The confirmation of the positivity was done by NAAT as recommended by CDC and WHO.7 Briefly, viral RNA was extracted using RNA extraction kit (Thermofisher Scientific) and tested immediately or stored at −70°C. Real-time fluorescent reverse-transcription polymerase chain reaction (RT-PCR) was performed to identify SARS-CoV-2 (BGI Biotechnology Co., Ltd, Wuhan, Hubei Province, PRC). The cycle threshold (Ct) value of the positive samples ranged from 15 to 35. Estimation of approximate viral load based on Ct values was done by performing RT-PCR in serially diluted FDA proficiency samples. The samples were serially diluted to produce final concentrations ranging from 1.8x107 to 1.8x102 and analyzed by RT-PCR in triplicate.
Antigen Tests
Antigen tests from seven IVD manufacturers were used to evaluate the analytical and clinical performance. These included: 1) BD Veritor™ System for rapid detection of SARS-CoV-2 (Becton, Dickinson and Company, Maryland, USA; Antigen kit #1), 2) CareStart™ COVID-19 Antigen (Accesas Bio, Inc., NJ, USA; Antigen kit #2), 3) SG Diagnostics Antigen detection kit (SG Diagnostics, Singapore; Antigen kit #3), 4) Sofia SARS Antigen FIA (Quedel Corporation, Hannover, Germany; Antigen kit #4), 5) Rapid Response™ COVID-19 Antigen Rapid Test (BNTX, Inc., ON, Canada; Antigen kit #5), 6) Shenzhen SARS-CoV-2 Antigen Test kit (Shenzhen Ultra-Diagnostics Biotec. Co., Ltd, Shenzhen, PRC; Antigen kit #6), and 7) Genedia W COVID-19 Ag (Green Cross Medical Sciences Corp, Chungcheongbuk, Republic of Korea; Antigen kit #7). Of these test kits, Antigen kit #1, #2, and #4 have been approved by the FDA under EUA. Antigen kit #4 was the first to receive the EUA and is based on lateral flow immunofluorescent sandwich assay. Specimen collection and testing were done using a method that we previously validated.12 Briefly, the pellet collected by high-speed centrifugation from 200 µL of UTM from each positive sample was reconstituted with extraction buffer provided by each antigen kit to disrupt viral particles and expose nucleocapsid proteins. Two to three drops (approx. 80 µL) of extracted swab sample is then applied to the test devices and the results were read and interpreted as per instruction manual of IVD manufacturers.
Evaluation of Analytical Performance
For analytical performance, heat-inactivated SARS-CoV-2 (ATCC® VR-1986HK™) was spiked to 500 µL of extraction fluid (assay buffer) provided by each test kit in order to give a final viral concentration of 4.6x104/mL, 9.5x104/mL, 1.85x105/mL, 3.75x105/mL, and 7.5x105/mL. Three drops (approx. 80 µL) of the spiked assay buffer were applied per test kit and the results were noted as recommended by the instruction manual of the IVD manufacturers. The approximate viral concentration per reaction were 3,750, 7,500, 15,000, 30,000, and 60,000, respectively.
Results and Discussion
The analytical performance of seven Ag-RDT kits were evaluated by analyzing five RT-PCR positive specimens. Surprisingly, all the specimens showed a negative result by all seven Ag-RDT except Antigen kit #6. When tested with Antigen kit #6, only one specimen gave a positive test result (Table 1). The Ct value of the specimen that showed the positive result was 15. All other specimens with a Ct value above 19 were negative for antigen testing. This indicates that antigen testing is far less sensitive than RT-PCR. To estimate the viral load from Ct value, we tested serially diluted FDA proficiency samples by RT-PCR. Limit of detection (LOD) of RT-PCR assay was 1.8x104 viral particles/mL which was detected in all triplicate runs with a Ct value ranging from 35–36 (Table 2). Ct values of 24–25 have an approximate viral concentration of 1.8x107/mL. As only one sample with a Ct value of 15–19 was positive for Ag-RDT, our result suggests that Ag-RDT are effective only when the viral load in the specimen is over 1.8x107/mL.
 |
Table 1 Comparison of Results of Seven Rapid Antigen Testing Kits with RT-PCR Positive Samples |
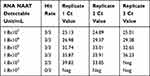 |
Table 2 Determination of Viral Load from RT-PCR Ct Value |
To assess the sensitivity of Ag-RDT, various concentrations of heat-inactivated SARS-CoV-2 were analyzed with antigen testing. Heat-inactivated SARS-CoV-2 were spiked into extraction buffer provided by IVD manufacturer to get a final concentration ranging from 4.6x104/mL to 7.5x105/mL. None of these samples were positive for all Ag-RDT (Table 3). This further supports that analytical sensitivity and LOD of Ag-RDT were poor and are suitable for use only when the viral load in the specimen is very high, as often seen in the patients with severe COVID-19 disease.
 |
Table 3 Comparison of Results of Seven Rapid Antigen Testing Kits with Various Concentrations of Heat-Inactivated SARS-CoV-2 |
Although Ag-RDT have diagnostic value comparable to antibody testing and are economic, rapid, and feasible for widespread testing in both laboratory and non-laboratory settings, it has poor clinical performance compared to the gold standard molecular assays. As global cases of COVID-19 continue to rise, it is expected that more IVD manufacturers will come up with Ag-RDT and more clinical laboratories will be using it to meet the testing demands. However, proper clinical evaluation of such antigen-based tests is crucial to add diagnostic value of Ag-RDT. Currently, there are a limited number of studies that provide the comparison of analytical and clinical sensitivity of Ag-RDT. Although done in small number of samples, our results of the comparison of seven Ag-RDT provides valuable information on its limitations and should help guide clinical laboratories and public health officials for the appropriate selection of diagnostic tests for COVID-19. Rapid immunochromatography-based tests have been widely used in clinical laboratories for the diagnosis of a wide variety of infectious diseases. The major problems with these immunoassay tests are, unlike PCR-based assay which allows amplification of targeted gene to greatly enhance its sensitivity, Ag-RDT are limited to the amount of antigens in the sample.
Although Ag-RDT has high specificity, many researchers have questioned its clinical utility because of its low sensitivity. Our result shows that Ag-RDT are unable to detect SARS-CoV-2 when the viral load in the sample is less than 1.8x107/mL and a Ct value more than 19. Systemic review on Ag-RDT showed that its clinical sensitivity is acceptable only when the viral loads are high with a Ct value below 25.13 Our result of low sensitivity of Ag-RDT is also supported by several other investigators. Liotti et al14 reported that the sensitivity of Ag-RDT compared to RT-PCR is over 95% when the Ct value is less than 25 but it declines drastically to 20–40% when the Ct value is greater than 25. Gannon et al,15,16 in their comparison study, found that Ag-RDT is 2–5-fold less sensitive compared to RT-PCR assay from respiratory specimens and their sensitivity varied from 22–70%. Furthermore, Lambert-Niclot et al17 also showed that overall sensitivity of Ag-RDT is only 50% when compared to RT-PCR, and Scohy et al18 reported that the sensitivity is only 32% and those that were detected had very high viral load. The poor clinical performance of Ag-RDT is because of its analytical sensitivity compared to molecular based assays. One of the major limitations of Ag-RDT is, unlike molecular assays, it does not offer the advantage of amplification of target RNA compromising sensitivity. However, it is generally believed that the specificity of Ag-RDT is high and comparable with molecular assays.19
It is now well evident that the severity of COVID-19 is directly dependent on the viral load.20 Therefore, perhaps one advantage of Ag-RDT can be useful in identifying and predicting the severity of disease. Since severity of the disease is often determined by massive proinflammatory response induced by SARS-CoV-2, measurement of proinflammatory cytokines (IL-6, IL-8, and TNF-a) together with Ag-RDT may aid diagnosis.21 On the other hand, the viral load in the sample peaks in 1–3 days of infection (Figure 1), therefore, collection of specimens at appropriate timing may increase the clinical sensitivity of Ag-RDT. Recent interim guidelines for antigen testing for SARS-CoV-2 formulated by CDC recommended a different strategy to interpret the result of antigen testing among symptomatic and asymptomatic individuals.19 In the symptomatic patients, a positive antigen test can be confirmatory while a negative result should be verified with NAAT. Whereas, in asymptomatic individuals, a negative antigen test result indicates no current evidence of infection and a positive result should be confirmed by NAAT.
The major limitation of this study is the small sample size. However, we have evaluated seven Ag-RDT that are available on the market. Despite the limitation of sample size, our study provides a fundamental basis to conduct a similar study on a large scale to validate the clinical utility of rapid antigen-based assays. Our future plan is to conduct the study with a large number of samples with all Ag-RDT assays available in the market for diagnostic use. Our current data suggests that none of these rapid tests are suitable to identify patients with low viral load. Therefore, Ag-RDT should not be the choice of test to identify asymptomatic patients with low viral load. Before implementing widespread Ag-RDT testing, more studies are needed to assess its clinical utility and diagnostic value.
Conclusion
Rapid antigen tests have low analytical and clinical sensitivity to identify asymptomatic patients with low viral load compared to molecular based assays. Therefore, analytical performance of rapid antigen tests should be thoroughly evaluated before implementing it at clinical decision level. More studies are needed to evaluate the clinical performance of antigen-based testing before implementing it for widespread testing.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Tang YW, Schmitz JE, Persing DH, Stratton CW. Laboratory diagnosis of COVID-19: current issues and challenges. J Clin Microbiol. 2020;58(6):e00512–20. doi:10.1128/JCM.00512-20
2. Li C, Zhao C, Bao J, Tang B, Wang Y, Gu B. Laboratory diagnosis of coronavirus disease-2019 (COVID-19). Clin Chim Acta. 2020;510:35–46. doi:10.1016/j.cca.2020.06.045
3. Russo A, Minichini C, Starace M, Astorri R, Calò F, Coppola N; Vanvitelli COVID-19 group. Current status of laboratory diagnosis for COVID-19: a narrative review. Infect Drug Resist. 2020;13:2657–2665. doi:10.2147/IDR.S264020
4. Bohn MK, Lippi G, Horvath A, et al. Molecular, serological, and biochemical diagnosis and monitoring of COVID-19: IFCC taskforce evaluation of the latest evidence. Clin Chem Lab Med. 2020;58(7):1037–1052. doi:10.1515/cclm-2020-0722
5. Khan S, Ali A, Shi H, et al. COVID-19: clinical aspects and therapeutics responses. Saudi Pharm J. 2020;28(8):1004–1008. doi:10.1016/j.jsps.2020.06.022
6. SARS-CoV-2 (COVID-19) testing guide. Available from: https://www.medicaldatabase.com/covid-19-testing-guide.
7. World Health Organization. Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases interim guidance; 2020. Available from: https://apps.who.int/iris/handle/10665/331329.
8. Mohammadi A, Esmaeilzadeh E, Li Y, Bosch RJ, Li JZ. SARS-CoV-2 detection in different respiratory sites: a systematic review and meta-analysis. E Bio Med. 2020;59:102903. doi:10.1016/j.ebiom.2020.102903
9. Chhikara BS, Rathi B, Singh J, Poonam. Corona virus SARS-CoV-2 disease COVID-19: infection, prevention and clinical advances of the prospective chemical drug therapeutics. Chem Biol Lett. 2020;7(1):
10. American Association of Clinical Chemistry. Coronavirus testing survey. Available from: https://www.aacc.org/science-and-research/covid-19-resources/aacc-covid-19-testing-survey.
11. World Health Organization. Antigen-detection in the diagnosis of SARS-CoV-2 infection using rapid immunoassays – interim guidance; 2020. Available from: https://www.who.int/publications/i/item/antigen-detection-in-the-diagnosis-of-sars-cov-2infection-using-rapid-immunoassays.
12. Beqaj S, Eshghifar N. Validation summary of UDX SARS-CoV-2 RT-PCR assay for detection of SARS-CoV-2 Coronavirus from saliva specimens. UltimateDx internal document; October 20, 2020. Available from https://www.fda.gov/media/137372/download.
13. Dinnes J, Deeks JJ, Adriano A, et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst Rev. 2020;8:CD013705. doi:10.1002/14651858.CD013705
14. Liotti FM, Menchinelli G, Lalle E, et al. Performance of a novel diagnostic assay for rapid SARS-CoV-2 antigen detection in nasopharynx samples. Clin Microbiol Infect. 2020. doi:10.1016/j.cmi.2020.09.030
15. Mak GCK, Lau SSY, Wong KKY, et al. Analytical sensitivity and clinical sensitivity of the three rapid antigen detection kits for detection of SARS-CoV-2 virus. J Clin Virol. 2020;133:104684. doi:10.1016/j.jcv.2020.104684
16. Mak GCK, Cheng PKC, Lau SSY, et al. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J Clin Virol. 2020;129:104500. doi:10.1016/j.jcv.2020.104500
17. Lambert-Niclot S, Cuffel A, Le Pape S, et al. Evaluation of a rapid diagnostic assay for detection of SARS-CoV-2 antigen in nasopharyngeal swabs. J Clin Microbiol. 2020;58(8):e00977–20. doi:10.1128/JCM.00977-20
18. Scohy A, Anantharajah A, Bodéus M, Kabamba-Mukadi B, Verroken A, Rodriguez-Villalobos H. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J Clin Virol. 2020;129:104455. doi:10.1016/j.jcv.2020.104455
19. Center for disease control and prevention: interim guidance for antigen testing for SARS-CoV-2. Dec 16, 2020 Available from https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antigen-tests-guidelines.html.
20. Imai M, Iwatsuki-Horimoto K, Hatta M, et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc Natl Acad Sci U S A. 2020;117:16587–16595.
21. Khatri M, Mago P. Nitazoxanide/Camostat combination for COVID-19: an unexplored potential therapy. Chem Biol Lett. 2020;7(3):
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.