Back to Journals » Infection and Drug Resistance » Volume 15
Evaluating the Role of Wastewaters as Reservoirs of Antibiotic-Resistant ESKAPEE Bacteria Using Phenotypic and Molecular Methods
Authors Mustafa SS, Batool R, Kamran M , Javed H, Jamil N
Received 23 April 2022
Accepted for publication 20 July 2022
Published 28 September 2022 Volume 2022:15 Pages 5715—5728
DOI https://doi.org/10.2147/IDR.S368886
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Syeda Samar Mustafa,1 Rida Batool,1 Muhammad Kamran,2 Hasnain Javed,3 Nazia Jamil1
1Institute of Microbiology and Molecular Genetics, Quaid e Azam Campus, University of the Punjab 54590, Lahore, Pakistan; 2Queensland Alliance for Agriculture and Food Innovation Centre for Animal Science, University of Queensland, Brisbane, Queensland, 4072, Australia; 3Provincial Public Health Reference Lab, Punjab AIDS Control Program, Lahore, Pakistan
Correspondence: Syeda Samar Mustafa, Email [email protected]
Introduction: Wastewaters carrying thousands of human specimens from the community and representing the diversity of antibiotic-resistant bacteria (ARB) and antibiotic-resistance genes (ARGs) directly from the community mirror the extent of AR spread in the community and environment. This study aimed to investigate the occurrence and distribution of antibiotic-resistant ESKAPEE bacteria in the community versus clinical settings through monitoring nonclinical and clinical wastewaters.
Methodology: Seven wastewater samples were collected from different environmental sources. Isolates were obtained on general and selective media, biochemically characterized and antimicrobial-susceptibility tests performed by disk diffusion against 13 antibiotics according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines using MastDisc disk cartridges, and 16S rRNA metagenomic analysis was performed for two water samples.
Results: Of 43 isolates, all representatives of the ESKAPEE group were recovered from clinical wastewaters, but Gram-positive cocci were not obtained from nonclinical wastewaters. The most predominant isolate was Pseudomonas aeruginosa (n=15; 33%), followed by Escherichia coli (n=9; 20%). Complete (100%) resistance to eleven of the tested antibiotics was observed, with only a few isolates being susceptible to clarithromycin, amikacin, and gentamicin. The lowest (79%) resistance rate was observed for linezolid. The multiple antibiotic resistance (MAR) index was calculated, and the resistance phenotype was independent of the wastewater source, indicated by x2 (P=0.766). Metagenomic analysis replicated the results, as Pseudomonas spp., Acinetobacter spp., and Escherichia spp. were found to be predominant. The integrase gene (IntI1) was also amplified in Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa.
Conclusion: Wastewaters are significant carriers of drug-resistant ESKAPEE bacteria and play an important role in their dissemination. This study endorses the periodic surveillance of water systems to evaluate the presence and burden of antibiotic-resistant pathogens.
Keywords: resistance profiling, ESKAPEE, wastewater, metagenome, IntI1
Graphical Abstract:
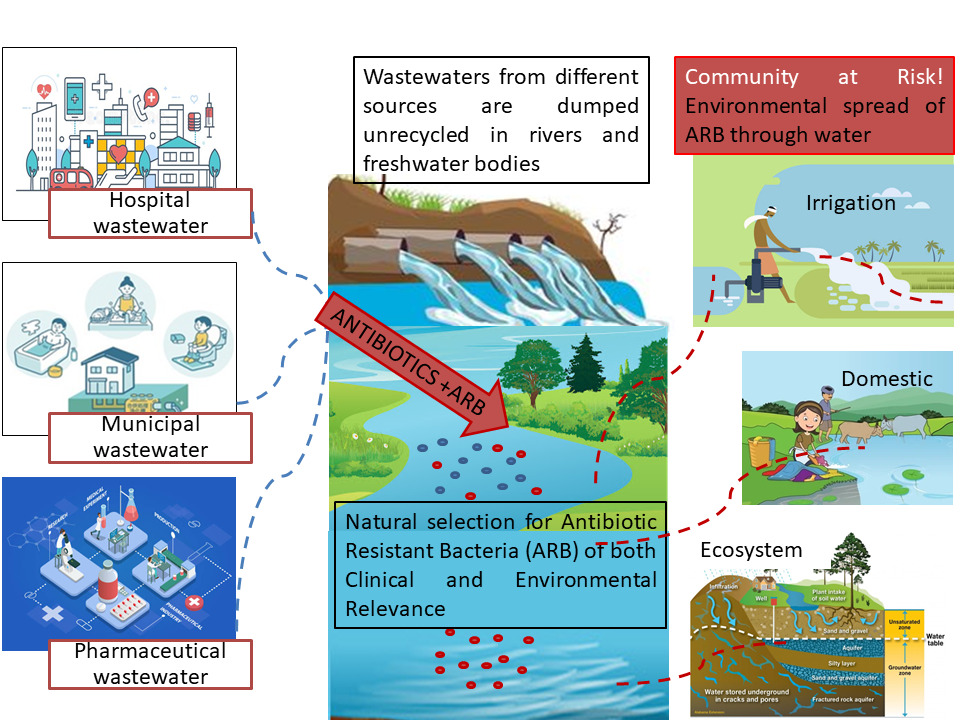
Introduction
Antimicrobial resistance has taken the form of a global pandemic in recent years and requires continuous and concerted efforts to mitigate its risks. Antibiotic-resistant bacteria (ARB) especially the multidrug resistant (MDR) ones, are widely spread in the environment.1 In 2017, a list of AR priority pathogens was published by World Health Organization (WHO), indicating the bacteria that pose a serious threat to mankind and to which new antibiotics are urgently needed. Depending on urgency, these bacteria have been given priority levels as critical, medium, and low.2–4 Most of these resistant pathogens include Gram-negative bacteria owing to their structure, which renders them more resistant than Gram-positive bacteria.4 Those at the critical level included Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacteriaceae (resistant to carbapenems and third-generation cephalosporins), while vancomycin-resistant enterococci (VRE) and methicillin-resistant Staphylococcus aureus were considered high priority ARB.2 ESKAPEE is an acronym for seven bacterial species — Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter spp., and Escherichia coli5 — that show high levels of antibiotic resistance. Overuse and misuse of antibiotics results in continual increase in antibiotic-resistant bacteria and environmental spread of antibiotic-resistance genes in environmental samples like wastewater of both clinical6–9 and nonclinical10–12 origin. Previous studies have shown that hospital wastewater acts as a selective environment for the development of ARB and then as a major contributor to increasing the burden of ARB in natural environments.8 Antibiotics exert a selection of ARB by killing or inhibiting the growth of susceptible bacteria. The huge amount of antibiotics used in hospitals and private households and their subsequent release into hospital and municipal wastes places a selective pressure on bacteria.13,14 Antibiotic-resistance genes, via horizontal gene transfer (HGT), are not only present in pathogens but also in environmental bacteria in terrestrial and aquatic environments,15 and they can be enriched through multiple human activities.16 Integrons are significant contributors to the acquisition and spread of ARGs, with the class 1 integron-integrase gene being the most important. The dissemination of this gene could be used as a proxy for anthropogenic contamination for a variety of reasons, including their association with antibiotic-resistance genes, their presence in pathogenic and nonpathogenic bacteria, their relative abundances being able to be altered through environmental factors, and playing a significant role in HGT, with the IntI1 genetic variant being present on diverse mobile genetic elements.17,18 Initially, class 1 integrons were reported from clinical isolates, but metagenomic and qPCR analyses have revealed their presence in diverse environments.19
Studies have shown the occurrence of ARB in foods and animals,20,21 and the incidence of ARB and antibiotic-resistance genes (ARGs) of clinical origin, ie, the contaminating resistome in the environment, signifies the increasing AR in humans.22 Some researchers look at the other side of the picture, ie, clinical AR is an endowment by environmental bacteria and ARGs. They propose that ARGs that have been presented by clinical bacteria evolve from natural antibiotic-producing microbes and their competitors in natural habitats.23,24 Antibiotic resistance in humans is either hospital-acquired or community-acquired, eg, MRSA and VRE MDR strains of E. coli and A. baumannii are increasingly being detected in community-acquired infections,22,25 though ARB occur commonly in hospitals, where they cause nosocomial infections.26,27 Of particular importance are ESKAPEE bacteria,28,29 which are largely environmental or commensal, cause opportunistic infections in immunocompromised people, and are not pathogenic otherwise. They have intrinsic resistance to one or more antibiotics and acquired resistance genes for additional antibiotics as well.30 Community risk for ARB can be effectively measured by analyzing wastewaters carrying thousands of human specimens from the community, which in turn can lead to the detection of new or rare ARB as well.31 Municipal wastewater-treatment plants are AMR hot spots, as they harbor numerous fecal and environmental bacteria under the influence of subinhibitory concentrations of antimicrobial compounds.32,33 In one study, a small number of wastewater samples were used to elucidate diversity in all urban runoffs, endorsing the applicability of a small sample as a screening tool for the existence of ARB in the community.31
In Pakistan and other developing countries, hospital wastewater is discharged into the drainage system and subsequently to lakes and river without any treatment.34 Few studies from Pakistan have been published on the role of wastewaters in the environmental spread of antibiotic resistance.6,35 This study, therefore, aims to investigate the occurrence and distribution of ARB in community versus clinical settings through monitoring nonclinical (municipal, pharmaceutical, and river) and clinical (hospital) wastewater and their ostensible role in the environmental spread of drug resistance via HGT by detection of the IntI1 integrase gene.
Methodology
Sample Collection
Seven wastewater samples from different environmental sources — hospital (3), municipal (2), pharmaceutical (1), and a river (1) — were collected over a period of 3 months. Hospital wastewater samples were collected from postsurgical, intensive care, and emergency wards. Municipal wastewater samples were collected from the drain-inflow site (where the sewer receives collected water) and near the settling tank (where water is stagnant). For each of the samples, 50 mL was collected in sterile plastic containers and transported within 2 hours of collection to the laboratory using a cooler box with ice packs. For metagenomic DNA isolation, 30 mL in triplicates for two water samples was collected.
Isolation and Biochemical Characterization of Bacterial Strains
All samples were simultaneously inoculated onto general-purpose (ie, nutrient agar) and selective media (ie, MaConkey and eosin–methylene blue (EMB) agar). Each sample was serially diluted up to a dilution factor of 10−5. Plates were analyzed for total bacterial count and colony morphology. Isolates were subjected to Gram-stain, motility, catalase (Figure S1), and oxidase tests for preliminary identification to differentiate between members of Enterobacteriaceae, P. aeruginosa, and A. baumannii. Gram-negative isolates were characterized using triple-sugar iron (TSI) and indole, methyl red, Voges–Proskauer, and citrate (IMViC) tests, while S. aureus was characterized by selective growth on mannitol salt agar (MSA) (Figure S2).
Antibiotic-Susceptibility Testing
Antibiotic-susceptibility testing (AST) was done for each of the bacterial isolates by disk diffusion (Figure S3). Cultures 24 hours old were transferred to nutrient broth (4 mL for each isolate) for overnight incubation at 37°C. Freshly grown cultures were standardized to turbidity of 0.5 McFarland standard. Bacterial lawns were prepared on Mueller–Hinton Agar, and antibiotic disks were placed aseptically onto the prepared plates. AST was performed against the following antibiotic disks (MastDiscs, Mast Group, Bootle, UK): Piperacillin (PRL, 30 µg), piperacillin–tazobactam (PTZ, 30/6 µg), cefuroxime (CXM, 5 µg), meropenem (MEM, 10 µg), ciprofloxacin (C, 5 µg), levofloxacin (LEV, 1 µg), amikacin (AMK, 30 µg), gentamicin (GM, 10 µg), vancomycin (VA, 5 µg), clarithromycin (CLA, 2 µg), azithromycin (ATH, 15 µg), clindamycin (CD 2 µg), and linezolid (LZD, 10 µg). E. coli ATCC 25922 and K. pneumoniae ATCC700603 were used as quality controls for disk diffusion testing. These strains were found to be susceptible to all antibiotics used. Diameters of zones of inhibition were measured after 24 hours of incubation and results interpreted according to EUCAST guidelines 2020.36 MDR strains were defined as resistant to one antibiotic from three or more antibiotic classes,37 while XDR (extremely drug-resistant) strains were those that were found to be susceptible to one or two classes of antibiotics.38 The multidrug-resistance index (MARI) was calculated as the ratio of the number of antibiotics to which the bacterial isolate was resistant to the total number of antibiotics included in susceptibility testing (Table S1).
Molecular Detection of IntI1 Gene
Bacterial strains were subjected to molecular detection of the IntI1 gene using IntI1-F TCTCGGGTAACATCAAGG and IntI1-R AGGAGATCCGAAGACCTC primers.39 For this purpose, DNA was isolated by boiling lysis with suspending loopful of overnight-grown pure bacterial culture in 300 µL sterile d.H2O and heating the Eppendorf tubes at 95°C for 10 minutes with intermittent vortexing. Tubes were centrifuged at 14,000 rpm for 1 minute to settle cell debris, and supernatant was shifted to a sterile tube. For PCR amplification, initial denaturation was done at 95°C for 5 minutes and subsequent final denaturation at 95°C for 45 seconds. Annealing was done at 55°C for 60 seconds. Initial extension was carried out at 72°C for 60 seconds and final extension done at 72°C for 10 minutes. The total number of cycles was 30. The amplified product was purified by using a GeneJET PCR purification kit (K0701). The purified product was sent for sequencing by Sanger dideoxy method to 1st Base Laboratories, Singapore. Sequencing was carried out using a BigDye Terminator version 3.1 cycle-sequencing kit using an Applied Biosystems thermocycler 9600. Sequenced files were analyzed using Chromas version 2.6.6 and sequences were submitted to the NCBI GenBank after similarity searches using the NCBI BLASTn database.
Metagenomic Analysis
Two of the water samples (ie, hospital wastewater and river water) were subjected to 16S ribosomal RNA metagenomic sequencing through the Illumina Miseq platform. A paired-end method was used to construct a fragment library for paired-end sequencing. The 16S V3–V4 region was amplified using the primer pair 5ʹACTCCTACGGGAGGCAGCAG3’ 5ʹGGACTACNNGGGTATCTAAT3ʹ.40 Raw paired-end reads (FASTQ) from the original DNA fragments were imported to Qiime2 version 2021.4 software.41 Paired-end reads for all samples were imported using the manifest file method. Quality filtering, denoising, and chimeric sequence removal were done using the DADA2 denoising method.42 To get constant read length for all reads, read truncation was used. A FASTA file with the sequences to use as reference with QIIME2 data-type feature data (sequence) was used. SILVA (https://www.arb-silva.de/download/archive/qiime) for 16S rRNA was used as a reference database. Naïve Bayes classifier and the q2-feature-classifier plugin43 were used to assign likely taxonomies to each read. This classifier was trained on the 97%-similarity OTU sequences from the reference database.
Statistical Analysis
Correlation of resistance status to the isolation source of the isolates was evaluated by x2 tests (Table S2 and S3), and percentage relative abundance of operational taxonomic units (OTUs) assigned to ESKAPEE genera (Table S4) was calculated using Microsoft Excel 2010.
Results
Seven wastewater samples from clinical and nonclinical sources were analyzed for the presence of drug-resistant isolates. A total of 92 isolates were randomly selected based on differences in colony morphology, of which 43 (clinical n=25; nonclinical n=18) were identified as ESKAPEE bacteria based on phenotypic and biochemical characteristics (Table S5). The distribution of bacteria among the wastewater samples showed Escherichia coli and Pseudomonas aeruginosa to be the most prominent isolates from hospital effluent, while Pseudomonas aeruginosa and Enterobacter spp. remained predominant in municipal wastewater. Pharmaceutical wastewater was the source of Klebsiella pneumoniae primarily. Gram-positive cocci were exclusively obtained from hospital wastewater, among which four isolates were identified as Staphylococcus aureus and two as enterococci. River water primarily contained Pseudomonas aeruginosa and Escherichia coli among the ESKAPEE bacteria (Figure 1). For each of the ESKAPEE isolates, there was no significant difference in numbers from clinical and nonclinical wastewaters, except for S. aureus and Enterococcus spp., which were recovered from clinical wastewater. Interestingly, all other bacteria were found to be more in number in nonclinical wastewaters (Figure 2).
Phenotypic Antibiotic-Susceptibility Testing
Susceptibility testing of the 43 isolated bacteria against different antibiotic classes exhibited widespread drug resistance, irrespective of wastewater source (Figure 3). Resistance patterns and MARI showed that most (n=30) isolates were XDRs, with being 100% resistance to all antibiotic categories tested against them. The rest (n=13) showed variable but large MARI (ranging from 0.7 to 0.9) indicating their MDR status (Table 1). Individual resistance phenotypes for each isolate indicated more XDR from both clinical and nonclinical wastewater sources (Table 2 and Figure 4). MDR was observed for four bacterial species: E. coli, K. pneumoniae, A. baumannii, and P. aeruginosa. The rest of the bacterial species were found to be XDR (Table S6).
 |
Table 2 Antibiogram of the isolated bacterial species from wastewater sources. Species-wise resistance rate against 13 antibiotics showed only a few isolates |
Metagenomics
Metagenomic analysis of samples revealed the presence of prominent members of the ESKAPEE group: Pseudomonas, Acinetobacter, and Escherichia spp. Their relative abundances somehow replicated the percentages of these isolates obtained via culturing. Just like phenotypic isolation, genotyping indicated Pseudomonas and Escherichia spp. as the most abundant in river water, while all Pseudomonas, Acinetobacter, and Escherichia spp. in hospital wastewater (Figure 5). Raw sequence reads have successfully been published with accession PRJNA797209 and SAMN24969811 and SAMN24969812 in SRA and Biosample NCBI databases, respectively.
Detection of IntI1 Gene
Nearly 42% (18 of 43) of the isolates were found to be positive for IntI1, among which a majority were E. coli, K. pneumoniae, and P. aeruginosa (Figure 6). Sequences for IntI1 integrase have been submitted to the NCBI GenBank and assigned accession numbers ON230319 and ON230320.
Statistical Analysis
Using x2 tests, drug-resistance status and wastewater source were found to be independent variables, ie, resistant strains were widespread throughout the water systems (P=0.766).
Discussion
In recent years, one of the most important pandemics is antimicrobial resistance in general and bacterial antibiotic resistance in particular after the emergence of MDR and XDR strains. In the present study, antibiotic-resistance patterns for ESKAPEE bacteria were analyzed, as the WHO has enlisted most of them as “priority level 1: critical group” bacteria. Wastewater is the most important vehicle for the environmental dissemination of drug-resistant bacteria and antibiotic-resistance genes (ARGs). Bacterial isolates from different sources were obtained and ESKAPEE bacteria identified by biochemical profiling. Of 92 isolates, 43 (46.7%) from wastewater sources belonged to the ESKAPEE group pointing to their spread in the environment. Of these 43 isolates, 25 (58.1%) were recovered from clinical wastewater sources and 18 (41.8%) from nonclinical wastewater. With respect to the total bacterial count, no significant difference was found among the samples. Such results are due to the fact that not only hospital wastewaters have high concentrations of antibiotics and disinfectants but also municipal, pharmaceutical, and industrial effluents carry heavy loads of antimicrobial compounds that allow resistant bacteria to outnumber nonresistant phenotypes in such environments. Widespread resistance to multiple antibiotics was also reported in a study from Bangladesh that compared the rate of resistance among ESKAPEE bacteria from clinical and nonclinical sources.9
The most frequent isolate was Pseudomonas aeruginosa (n=15) in both clinical and nonclinical effluent, followed by Escherichia coli (n=9). This frequency can be attributed to the extraordinary potential of Pseudomonas aeruginosa and E. coli in biofilm formation in sewerage systems, as many studies have correlated the extent of biofilm formation and drug resistance in isolates from wastewaters.44,45 Several studies have reported these Gram-negative rods as predominant drug-resistant species in hospital runoffs in addition to Klebsiella and Enterobacter spp.46–48 All of the seven ESKAPEE representatives were recovered from hospital wastewater, but S. aureus and enterococci were not obtained from nonclinical wastewaters. Somewhat similar results were reported in a study where the percentage of MRSA and VRE in different wastewater samples was found to be lowest compared to Gram-negative bacteria from the ESKAPEE group, but in contrast to the present study, they recovered very low methicillin-resistant S. aureus (MRSA) (37.5%) from hospital effluent compared to municipal wastewater (75%) and no vancomycin-resistant Enterococci (VRE) from hospital water (89.3%).12
Pan-drug resistance (PDR) refers to resistance of the isolate to all the compounds in all the categories of antibiotics, while some studies use PDR and XDR synonymously.38,49,50 However, for the current study, isolates showing 100% resistance to all included drugs were considered XDR to overcome study bias, as AST was performed only for the available classes of antibiotics. Overall, XDR phenotypes were commonly observed irrespective of the wastewater source: 73.9% (clinical) and 72.2% (nonclinical) mainly showing susceptibility toward a few antibiotics, including AMK, GM, and LZD. Resistance to gentamicin was also found to be uncommon (3.5%) in another study carried out on E. coli from hospital and community wastewaters in Norway.31 Percentages of MDR phenotypes in clinical and nonclinical sources were 34.7% and 27.7%, respectively (Figure 3). The results were validated with x2 tests that indicated no statistical significance between the two variables, ie, resistance phenotype and wastewater source (P=0.766). Further MDR strains were observed for E. coli, P. aeruginosa, A. baumannii, and K. pneumoniae, while the three other species were almost exclusively found to be XDRs (Figure 4). Several studies have reported MDR Gram-negative bacteria from environmental sources where their resistance pattern was established using disk diffusion according to either EUCAST51,52 or Clinical and Laboratory Standards Institute (CLSI) guidelines.53–56 The high incidence of extensively drug-resistant strains in both clinical and nonclinical wastewaters may be attributed to the constant selective pressure placed on the bacteria not only by the excessive use of antibiotics but also by other antimicrobial agents that are released from hospitals and other urban runoffs, eg, industrial and agricultural effluent. In Pakistan, the situation is worsened by the collection of hospital effluent into main drains, eg, wastewater from Jinnah Hospital is dumped into the Sattukatla drain, while the Shadman drain receives effluent from the Services Institute of Medical Sciences (SIMS) and Mayo Hospital.6 Such mixing of wastewaters promotes the selection of more resistant strains of diverse bacterial species by putting overwhelming selective pressure on antimicrobials. All these main sewers fall into the river Ravi without any treatment. One substantial contributor to nonclinical wastewater is pharmaceutical effluent,57 and this may favor the selection of resistant bacteria, among which the fitter ones are XDR strains. All isolates were found to be completely resistant to the following categories of antibiotics: β-lactams, including penicillin (PRL and PTZ), cephalosporin (CXM), and carbapenem (MEM); fluoroquinolones, including ciprofloxacin (C) and levofloxacin (LEV); macrolides, including azithromycin (ATH) and clindamycin (CD); and glycopeptides, including vancomycin (VA). A study conducted on wastewaters in Pakistan also reported high rates of resistance (58%–89%) for all the tested β-tactams, while high MIC values were observed for ciprofloxacin (256 µg/mL) and Gram-negative bacteria, including E. coli (28%) and Pseudomonas spp. (7%),35 among ESKAPEE members. A few isolates of P. aeruginosa and A. baumannii showed susceptibility to clarithromycin (macrolide). Most bacterial species showed susceptibility towards aminoglycosides, including amikacin (AK), and gentamicin (GM) and oxazolidinone, including linezolid (LZD) (Table 2). Sensitivity towards aminoglycosides has been reported from isolates of wastewater systems in Ethiopia.33
Both of the Gram-positive species were XDR, with some strains being susceptible to just linezolid. Among Gram-negative bacteria, Enterobacter spp. from both hospital and nonhospital wastewater were found to be exclusively XDR. Another study reported Enterobacter spp. were resistant to all the 12 antibiotics tested against them, while Klebsiella and Pseudomonas spp. followed, showing resistance to 11 and 10 antibiotics, respectively.48
Metagenomic analysis was done to evaluate the extent of spread of ESKAPEE bacteria in freshwater bodies through wastewater contamination by selecting two termini of the wastewater-collection system. Impact of anthropogenic activities on river-water quality and microbiome was reported in a study from Malaysia, where 100% of E. coli, K. pneumoniae, and E. cloacae and nearly 85% of E. faecalis were found to be MDR in the river Melayu, indicating the presence of MDR ESKAPEE members in water systems.55 Persistence of most important opportunistic pathogens that bear both environmental and clinical significance like Escherichia spp., Pseudomonas spp., and Acinetobacter spp., may clearly be attributed to their structural and functional virulence factors9,57 that lead to their selection through the course of the water-collection system and final dumping into the river. Integrons are classified as classes 1, 2, and 3 based on the integrase gene (intI1), with class 1 being the first detected and most frequent type in antibiotic-resistant bacteria.5 They can easily attain many genes to acquire resistance in bacteria. This study showed the widespread drug resistance among ESKAPEE through AST and that they are involved in HGT of drug-resistance genes as indicated by the presence of IntI1 gene, so we can say that these bacteria are important contributors to environmental dissemination of drug resistance. Class 1 integron (intI1) genes were found to be abundantly present in a wastewater-treatment plant in Texas in addition to the presence of MDR E. coli.54 A positive-feedback loop is established for long-term sustenance of ARB if such contaminated runoffs are used for agricultural, industrial, or domestic purposes, as in third-world countries. Most significantly, the presence of XDRs in both clinical and nonclinical wastewaters indicate a constant pressure that has resulted from indiscriminate use of antibiotics and other antimicrobials and their subsequent release into the environment, paving a way to the development and/or selection of MDR and XDR strains among bacterial species. This study provides a dimension to check the abundance of pathogenic bacteria in water systems, and such studies must be conducted periodically for the environmental surveillance of ARB and ARGs, and the factors that result in their enrichment in any given environment must be evaluated subsequently.
Data Sharing
Data sets related to the current study can be found in Tables S1–S6 and Figures S1–S3.
Ethics Approval and Consent to Participate
Not applicable, as the study did not include any human participants, human data, or human tissue.
Acknowledgments
Funding was provided by a University of the Punjab research grant for the fiscal year 2021–2022. The work presented in this study is part of the PhD thesis of the first author, Ms Syeda Samar Mustafa.
Author Contributions
All authors made a significant contribution to the work reported, whether in the conception, study design, execution, acquisition of data, analysis and interpretation, or all these areas, took part in drafting, revising, or critically reviewing the article, gave final approval to the version to be published, have agreed on the journal to which the article has been submitted, and agree to be accountable for all aspects of the work.
Funding
No formal funding or grant was provided for this study.
Disclosure
The authors declare that they have no conflicts of interest in relation to this work.
References
1. Kraemer SA, Ramachandran A, Perron GG. Antibiotic pollution in the environment: from microbial ecology to public policy. Microorganisms. 2019;7(6):180. doi:10.3390/microorganisms7060180
2. Tacconelli E, Carrara E, Savoldi A, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318–327. doi:10.1016/S1473-3099(17)30753-3
3. Havenga B, Ndlovu T, Clements T, Reyneke B, Waso M, Khan W. Exploring the antimicrobial resistance profiles of WHO critical priority list bacterial strains. BMC Microbiol. 2019;19(1):1–16. doi:10.1186/s12866-019-1687-0
4. Breijyeh Z, Jubeh B, Karaman R. Resistance of Gram-negative bacteria to current antibacterial agents and approaches to resolve it. Molecules. 2020;25(6):1340. doi:10.3390/molecules25061340
5. Partridge SR, Kwong SM, Firth N, Jensen SO. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev. 2018;31(4):e00088–00017. doi:10.1128/CMR.00088-17
6. Ahmad M, Khan AU, Wahid A, Butt ZA, Farhan M, Ahmad F. Role of hospital effluents in the contribution of antibiotics and antibiotic resistant bacteria to the aquatic environment. Pak J Nutr. 2012;11(12):1177. doi:10.3923/pjn.2012.1177.1182
7. Pandey R, Mishra SK, Shrestha A. Characterisation of ESKAPE pathogens with special reference to multidrug resistance and biofilm production in a nepalese hospital. Infect Drug Resist. 2021;14:2201. doi:10.2147/IDR.S306688
8. Yang C, Lin M, Liao P, et al. Comparison of antimicrobial resistance patterns between clinical and sewage isolates in a regional hospital in Taiwan. Lett Appl Microbiol. 2009;48(5):560–565. doi:10.1111/j.1472-765X.2009.02572.x
9. Rabbani M, Howlader MZH, Kabir Y. Detection of multidrug resistant (MDR) bacteria in untreated waste water disposals of hospitals in Dhaka City, Bangladesh. J Glob Antimicrob Resist. 2017;10:120–125. doi:10.1016/j.jgar.2017.04.009
10. Savin M, Bierbaum G, Hammerl JA, et al. ESKAPE bacteria and extended-spectrum-β-lactamase-producing Escherichia coli isolated from wastewater and process water from German poultry slaughterhouses. Appl Environ Microbiol. 2020;86(8):e02748–e02719. doi:10.1128/AEM.02748-19
11. Giebułtowicz J, Tyski S, Wolinowska R, et al. Occurrence of antimicrobial agents, drug-resistant bacteria, and genes in the sewage-impacted Vistula River (Poland). Environ Sci Pollut Res. 2018;25(6):5788–5807. doi:10.1007/s11356-017-0861-x
12. Nishiyama M, Praise S, Tsurumaki K, Baba H, Kanamori H, Watanabe T. Prevalence of antibiotic-resistant bacteria ESKAPE among healthy people estimated by monitoring of municipal wastewater. Antibiotics. 2021;10(5):495. doi:10.3390/antibiotics10050495
13. Kruse H. Indirect transfer of antibiotic resistance genes to man. Acta Vet Scand Suppl. 1999;92:59–65.
14. He Y, Yuan Q, Mathieu J, et al. Antibiotic resistance genes from livestock waste: occurrence, dissemination, and treatment. NPJ Clean Water. 2020;3(1):1–11. doi:10.1038/s41545-020-0051-0
15. Bello-López JM, Cabrero-Martínez OA, Ibáñez-Cervantes G, et al. Horizontal gene transfer and its association with antibiotic resistance in the genus Aeromonas spp. Microorganisms. 2019;7(9):363. doi:10.3390/microorganisms7090363
16. Adelowo OO, Helbig T, Knecht C, Reincke F, Mäusezahl I, Müller JA. High abundances of class 1 integrase and sulfonamide resistance genes, and characterisation of class 1 integron gene cassettes in four urban wetlands in Nigeria. PLoS One. 2018;13(11):e0208269. doi:10.1371/journal.pone.0208269
17. Gillings MR, Gaze WH, Pruden A, Smalla K, Tiedje JM, Zhu Y-G. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 2015;9(6):1269–1279. doi:10.1038/ismej.2014.226
18. Zheng W, Huyan J, Tian Z, Zhang Y, Wen X. Clinical class 1 integron-integrase gene–A promising indicator to monitor the abundance and elimination of antibiotic resistance genes in an urban wastewater treatment plant. Environ Int. 2020;135:105372. doi:10.1016/j.envint.2019.105372
19. Ma L, Li A-D, Yin X-L, Zhang T. The prevalence of integrons as the carrier of antibiotic resistance genes in natural and man-made environments. Environ Sci Technol. 2017;51(10):5721–5728. doi:10.1021/acs.est.6b05887
20. Founou LL, Founou RC, Essack SY. Antibiotic resistance in the food chain: a developing country-perspective. Front Microbiol. 2016;7:1881. doi:10.3389/fmicb.2016.01881
21. Perreten V, Schwarz F, Cresta L, Boeglin M, Dasen G, Teuber M. Antibiotic resistance spread in food. Nature. 1997;389(6653):801–802. doi:10.1038/39767
22. Tacconelli E, Sifakis F, Harbarth S, et al. Surveillance for control of antimicrobial resistance. Lancet Infect Dis. 2018;18(3):e99–e106. doi:10.1016/S1473-3099(17)30485-1
23. Arshad M, Zafar R. Antibiotics, AMRs, and ARGs: fate in the environment. In: Antibiotics and Antimicrobial Resistance Genes in the Environment. Elsevier; 2020:138–154.
24. Aminov RI, Mackie RI. Evolution and ecology of antibiotic resistance genes. FEMS Microbiol Lett. 2007;271(2):147–161. doi:10.1111/j.1574-6968.2007.00757.x
25. Gottlieb T, Nimmo GR. Antibiotic resistance is an emerging threat to public health: an urgent call to action at the Antimicrobial Resistance Summit 2011. Med J Aust. 2011;194(6):281–283. doi:10.5694/j.1326-5377.2011.tb02973.x
26. Hocquet D, Muller A, Bertrand X. What happens in hospitals does not stay in hospitals: antibiotic-resistant bacteria in hospital wastewater systems. J Hosp Infect. 2016;93(4):395–402. doi:10.1016/j.jhin.2016.01.010
27. Santajit S, Indrawattana N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed Res Int. 2016;2016:1–8. doi:10.1155/2016/2475067
28. Slavcovici A, Maier C, Radulescu A. Antimicrobial resistance of ESKAPE-pathogens in culture-positive pneumonia. Farmacia. 2015;63(2):201–205.
29. Navidinia M. The clinical importance of emerging ESKAPE pathogens in nosocomial infections; 2016.
30. Wyres KL, Holt KE. Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr Opin Microbiol. 2018;45:131–139. doi:10.1016/j.mib.2018.04.004
31. Paulshus E, Kühn I, Möllby R, et al. Diversity and antibiotic resistance among Escherichia coli populations in hospital and community wastewater compared to wastewater at the receiving urban treatment plant. Water Res. 2019;161:232–241. doi:10.1016/j.watres.2019.05.102
32. Kaplan E, Sela N, Doron-Faigenboim A, Navon-Venezia S, Jurkevitch E, Cytryn E. Genomic and functional characterization of qnr-encoding plasmids from municipal wastewater biosolid Klebsiella pneumoniae isolates. Front Microbiol. 2015;6:1354. doi:10.3389/fmicb.2015.01354
33. Teshome A, Alemayehu T, Deriba W, Ayele Y. Antibiotic resistance profile of bacteria isolated from wastewater systems in Eastern Ethiopia. J Environ Public Health. 2020;2020:1–10. doi:10.1155/2020/2796365
34. Al Aukidy M, Al Chalabi S, Verlicchi P. Hospital wastewater treatments adopted in Asia, Africa, and Australia. In: Verlicchi P, editor. Hospital Wastewaters: Characteristics, Management, Treatment and Environmental Risks. Cham: Springer International Publishing; 2018:171–188.
35. Saima S, Fiaz M, Manzoor M, et al. Molecular investigation of antibiotic resistant bacterial strains isolated from wastewater streams in Pakistan. Biotech. 2020;10(9):1–11. doi:10.3390/biotech10010001
36. The European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and Zone diameters; 2020. Available from: http://www.eucast.org.
37. Magiorakos A-P, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi:10.1111/j.1469-0691.2011.03570.x
38. Zhi-Wen Y, Yan-Li Z, Man Y, Wei-Jun F. Clinical treatment of pandrug-resistant bacterial infection consulted by clinical pharmacist. Saudi Pharm J. 2015;23(4):377–380. doi:10.1016/j.jsps.2015.01.001
39. Firoozeh F, Mahluji Z, Khorshidi A, Zibaei M. Molecular characterization of class 1, 2 and 3 integrons in clinical multi-drug resistant Klebsiella pneumoniae isolates. Antimicrob Resist Infect Control. 2019;8(1):1–7. doi:10.1186/s13756-019-0509-3
40. Tawfik SA, Azab MM, Ahmed AAA, Fayyad DM. Illumina MiSeq sequencing for preliminary analysis of microbiome causing primary endodontic infections in Egypt. Int J Microbiol. 2018;2018:1–15. doi:10.1155/2018/2837328
41. Estaki M, Jiang L, Bokulich NA, et al. QIIME 2 enables comprehensive end‐to‐end analysis of diverse microbiome data and comparative studies with publicly available data. Curr Protoc Bioinformatics. 2020;70(1):e100. doi:10.1002/cpbi.100
42. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581–583. doi:10.1038/nmeth.3869
43. Bokulich NA, Kaehler BD, Rideout JR, et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2ʹs q2-feature-classifier plugin. Microbiome. 2018;6(1):1–17. doi:10.1186/s40168-018-0470-z
44. Auguet O, Pijuan M, Borrego CM, et al. Sewers as potential reservoirs of antibiotic resistance. Sci Total Environ. 2017;605:1047–1054. doi:10.1016/j.scitotenv.2017.06.153
45. Nain Z, Islam M, Minnatul Karim M. Antibiotic resistance profiling and molecular phylogeny of biofilm forming bacteria from clinical and non-clinical environment in southern part of Bangladesh. Int J Enteric Pathog. 2019;7(2):37–43. doi:10.15171/ijep.2019.10
46. Ekhaise F, Omavwoya B. Influence of hospital wastewater discharged from University of Benin Teaching Hospital (UBTH), Benin City on its receiving environment. Am-Eurasian J Agric Environ Sci. 2008;4(4):484–488.
47. Chagas TPG, Seki L, Cury J, et al. Multiresistance, beta‐lactamase‐encoding genes and bacterial diversity in hospital wastewater in Rio de Janeiro, Brazil. J Appl Microbiol. 2011;111(3):572–581. doi:10.1111/j.1365-2672.2011.05072.x
48. Moges F, Endris M, Belyhun Y, Worku W. Isolation and characterization of multiple drug resistance bacterial pathogens from waste water in hospital and non-hospital environments, Northwest Ethiopia. BMC Res Notes. 2014;7(1):1–6. doi:10.1186/1756-0500-7-215
49. Pontikis K, Karaiskos I, Bastani S, et al. Outcomes of critically ill intensive care unit patients treated with fosfomycin for infections due to pandrug-resistant and extensively drug-resistant carbapenemase-producing Gram-negative bacteria. Int J Antimicrob Agents. 2014;43(1):52–59. doi:10.1016/j.ijantimicag.2013.09.010
50. Alkofide H, Alhammad AM, Alruwaili A, et al. Multidrug-resistant and extensively drug-resistant enterobacteriaceae: prevalence, treatments, and outcomes–a retrospective cohort study. Infect Drug Resist. 2020;13:4653. doi:10.2147/IDR.S283488
51. Dimitrova L, Kaleva M, Zaharieva MM, et al. Prevalence of antibiotic-resistant Escherichia coli isolated from swine faeces and Lagoons in Bulgaria. Antibiotics. 2021;10(8):940. doi:10.3390/antibiotics10080940
52. Roulová N, Mot’ková P, Brožková I, Pejchalová M. Antibiotic resistance of Pseudomonas aeruginosa isolated from hospital wastewater in the Czech Republic. J Water Health. 2022;20(4):692–701. doi:10.2166/wh.2022.101
53. Ohene Larbi R, Ofori LA, Sylverken AA, Ayim-Akonor M, Obiri-Danso K. Antimicrobial resistance of Escherichia coli from broilers, pigs, and cattle in the greater kumasi metropolis, Ghana. Int J Microbiol. 2021;2021. doi:10.1155/2021/5158185
54. Mukherjee M, Laird E, Gentry TJ, Brooks JP, Karthikeyan R. Increased antimicrobial and multidrug resistance downstream of wastewater treatment plants in an urban watershed. Front Microbiol. 2021;12:657353. doi:10.3389/fmicb.2021.657353
55. Ho JY, Jong M-C, Acharya K, et al. Multidrug-resistant bacteria and microbial communities in a river estuary with fragmented suburban waste management. J Hazard Mater. 2021;405:124687. doi:10.1016/j.jhazmat.2020.124687
56. Gumede SN, Abia AL, Amoako DG, Essack SY. Analysis of wastewater reveals the spread of diverse extended-spectrum β-lactamase-producing E. coli strains in uMgungundlovu District, South Africa. Antibiotics. 2021;10(7):860. doi:10.3390/antibiotics10070860
57. Guardabassi L, Petersen A, Olsen JE, Dalsgaard A. Antibiotic resistance in Acinetobacter spp. isolated from sewers receiving waste effluent from a hospital and a pharmaceutical plant. Appl Environ Microbiol. 1998;64(9):3499–3502. doi:10.1128/AEM.64.9.3499-3502.1998
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.







