Back to Journals » Cancer Management and Research » Volume 11
ERRα is an aggressive factor in lung adenocarcinoma indicating poor prognostic outcomes
Authors Li P, Wang J, Wu D, Ren X, Wu W, Zuo R, Zeng Q , Wang B, He X , Yuan J, Xie N
Received 10 February 2019
Accepted for publication 28 July 2019
Published 2 September 2019 Volume 2019:11 Pages 8111—8123
DOI https://doi.org/10.2147/CMAR.S204732
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Ping Li,1–3,* Jian Wang,4,* Desheng Wu,3 Xiaohu Ren,3 Wen Wu,2,3 Ran Zuo,2 Qingbo Zeng,3 Bingyu Wang,2 Xi He,1 Jianhui Yuan,2,5 Ni Xie1,2
1Biobank, Shenzhen Second People’s Hospital, First Affiliated Hospital of Shenzhen University, Shenzhen 518035, People’s Republic of China; 2Department of Medicine, University of South China, Hengyang 421001, People’s Republic of China; 3Department of Toxicology, Shenzhen Center for Disease Control and Prevention, Shenzhen 518035, People’s Republic of China; 4Department of Thoracic Surgery, The Shenzhen People’s Hospital, Shenzhen 518020, People’s Republic of China; 5Department of Occupational Health, Shenzhen Nanshan District Center for Disease Control and Prevention, Shenzhen 518054, People’s Republic of China
Correspondence: Ni Xie
Biobank Shenzhen Second People’s Hospital, First Affiliated Hospital of Shenzhen University, No. 3002 Sungang West Road, Futian District, Shenzhen 518035, People’s Republic of China
Tel +86 1 350 158 0802
Fax +86 7 558 300 3435
Email [email protected]
Jianhui Yuan
Department of Occupational Health, Shenzhen Nanshan District Center for Disease Control and Prevention, No. 95 Nanshang Road, Nanshan District, Shenzhen 518054, People’s Republic of China
Tel +86 1 350 158 0509
Fax +86 7 558 300 3435
Email [email protected]
*These authors contributed equally to this work
Purpose: Lung cancer is one of the most life-threatening cancer worldwide with poor prognosis attributed to the lack of early diagnosis and proper therapy. The estrogen-related receptor alpha (ERRα) is a multifunctional protein not limited to bind ligands and has been reported to be associated with numerous cancers. This study aimed to investigate the potential role of ERRα in lung cancer and to provide a novel perspective for lung cancer early diagnosis, targeted therapy, and prognosis assessment.
Methods: The correlation between ERRα mRNA expression and survival time of the online clinical data about lung cancer was analyzed by using Kaplan–Meier (KM) plotter. A mouse model of lung adenocarcinoma (LUAD) was constructed to detect the expression level of ERRα in tumor tissues. ERRα-knockdown LUAD cells were generated and the impacts of ERRα on cell proliferation, invasion, and metastasis were further analyzed. Cancerous and paracancerous tissues were collected to semi-quantitative the levels of ERRα in LUAD clinical samples (n=88), combined with clinical information for prognostic analysis.
Results: The KM plotter analysis suggested that ERRα is correlated with poor prognosis in LUAD (n=720) rather than in lung squamous cell carcinoma (LSCC) (n=524). ERRα is also upregulated in tumor tissues obtained from LUAD model mice. Quantitative analysis suggested an abnormal elevation of ERRα in LUAD cells rather than in LSCC cells. The results demonstrated that downregulation of ERRα impairs proliferation, invasion and migration abilities (P<0.01). The prognostic analysis showed that the overexpressed ERRα in LUAD was positively correlated with low survival rates (HR=1.597). The results indicate that the death risk of ERRα high expression is 1.597 times higher than ERRα low level in LUAD patients.
Conclusion: In summary, our findings suggest that ERRα is a potential aggressive factor of LUAD which implies poor prognosis.
Keywords: lung adenocarcinoma, ERRα, estrogen-related receptor alpha, proliferation, migration, metastasis, poor prognosis
Introduction
Lung cancer is one of the cancers with highest mortality rate worldwide.1 Non-small cell lung cancer (NSCLC) accounts for 75–80% of the total cases of lung cancers. The three main subtypes of NSCLC are lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LSCC), and large-cell carcinoma.2 LUAD is an important subtype of NSCLC. At present, the incidence of LUAD has surpassed that of LSCC, and LUAD has become the most common histological subtype of lung cancer.3 The pathogenesis of LUAD and related mechanisms still remain largely unknown. In our previous studies, we identified the abnormal elevation of ERRα in LUAD cells (A549, H1975, H1395) and mouse model. However, the role of ERRα in LUAD still needs further elucidation.
ERRα is one of the orphan nuclear receptors which can produce biological functions without binding to a ligand.4 By using the cDNA of the DNA-binding domain of estrogen receptor α (ERα) as a probe, ERRα was first screened by Giguere et al.5 ERRα not only participates in and affects the estrogen receptor signaling system but also participates in many metabolic processes such as glucose metabolism, lipid metabolism, and mitochondrial oxidative metabolism.6–9 ERRα was also found associated with the occurrence of metabolic diseases, such as obesity, diabetes, and osteoporosis.10–12 In recent years, studies have found that the expression of ERRα is closely related to estrogen-dependent tumors such as breast cancer, prostate cancer, and cervical cancer, as well as non-estrogen-dependent tumors such as gastric adenocarcinoma and colorectal cancer, which suggest that ERRα is involved in the process of tumor development.6,13–15 It has also been found that the expression of ERRα was upregulated in LUAD cell line A549, which promoted the proliferation of A549 cells in vitro.16 However, the role of ERRα in LUAD has not yet been fully understood. To further elucidate the function of ERRα, we established ERRα-knockdown LUAD cells (A549-ERRα-ko, H1975-ERRα-ko, H1395-ERRα-ko). Then, multiple malignant properties in foregoing cell models such as proliferation, invasion, and migration were investigated by CCK8 assay, Transwell migration assay, and scratch wound healing assay, respectively. The cell cycle was also measured by flow cytometry analysis. Moreover, we evaluated expression levels of ERRα in clinical samples (adjacent/cancerous tissues) by immunohistological staining. The association between ERRα and prognosis of LUAD was also analyzed.
Materials and methods
Database analysis
The relationship between ERRα mRNA level and survival rate in 720 patients with LUAD (n=720) was analyzed by using an online prognostic analysis tool Kaplan–Meier plotter (http://kmplot.com/analysis/). The relationship between ERRα mRNA level and survival rate in 524 patients with LSCC was also analyzed (n=524). Overall survival (OS) was chosen for evaluating patient’s survival. The correlation between patient’s survival rate and the foregoing two lung cancer subtypes was analyzed separately. The background database is manually curated. Gene expression data, relapse-free information, and overall survival information were downloaded from GEO (Affymetrix microarrays only), EGA, and TCGA. To analyze the potential role of ERRα in lung cancer, the patient samples were split into two groups according to the expression level of ERRα. The two patient cohorts were compared by a Kaplan–Meier survival plot, and the hazard ratio with 95% confidence intervals and log-rank P-value is calculated.
Cell culture
LUAD cells (A549), LSCC cells (SW-900, NCI-H520), and bronchial epithelial BEAS-2B cells were purchased from the American Type Culture Collection (ATCC). NCI-H1395 and NCI-H1975 (LUAD cells) were purchased from Stem Cell Bank, Chinese Academy of Sciences. Cells were cultured in RPMI 1640 or DMEM medium (Gibco) supplemented with 10% fetal bovine serum at 37°C in a humidified 5% CO2 atmosphere. Cells in logarithmic growth phase were used for experiments performed in this study.
Collection of clinical samples
A total of 92 cases of cancer tissue and 88 cases of paracancerous tissues were covered in a lung cancer tissue chip (Shanghai Outdo Biotech Company). The tissues on the chip were obtained from patients with lung cancer who underwent surgery from January 2008 to July 2013 and were followed-up for 3–8 years.
Quantitative real-time PCR
Total RNA was isolated using the RNAiso Plus reagent (Takara, Japan). cDNA was synthesized through reverse transcription using a 10 μL system of Reverse Transcript Kit (PrimeScript RT Master Mix) prepared on ice. Quantitative Real-Time PCR was performed on a Real-Time PCR System (ABI 7500, USA) using validated primers and SYBR Premix Ex Taq II (Takara, Japan). The number of cycle when the fluorescence first reached a preset threshold (Ct) was used to quantify the initial concentration of individual templates. The resulting expression of ERRα in each sample was normalized to the corresponding expression of internal control β-actin. qPCR primer pairs were as follows:
ERRα,
Forward: 5ʹ-GTCCAAAGGGTTCCTCGGAG-3ʹ
Reverse: 5ʹ-GGATGCCACACCATAGTGGTA-3ʹ;
β-actin,
Forward: 5ʹ-CATGTACGTTGCTATCCAGGC-3ʹ
Reverse: 5ʹ-CTCCTTAATGTCACGCACGAT-3ʹ.
Knockdown of ERRα in LUAD cells
The ERRα gene silencing sequence was ligated into the vector plvx-shRNA2p to construct the ERRα gene-silencing shRNA lentiviral vector. The shRNA with the best interference efficiency against ERRα gene was screened before the corresponding lentivirus was packaged. Lentivirus was used to transfect A549, H1395, and H1975 cells. The successfully transfected cells were screened by culture medium containing puromycin.
CCK8 viability assay
Cell viability was determined by Cell Counting Kit-8 (Dojindo, Laboratories, Shanghai, China). ERRα-knockdown LUAD cells (A549, NCI-H1395, and NCI-H1975) or LUAD cells transfected with empty vector were seeded into a 96-well plate (at density of 2000 cells/well) and placed in a cell culture incubator until the cells adhered to the bottom of the well. Serum-free medium containing 10% CCK-8 solution was added to each well at 24, 48, and 72 hrs, followed by incubation at 37°C for 3 hrs. The absorbance value of each well was read at 450 nm using a microplate reader (Tecan M1000, Männedorf, Switzerland). The experiments were repeated 3 times.
Cell cycle
Cell cycle changes were detected using the Cell cycle staining Kit (MultiSciences). Cells were suspended and adjusted to a density of 2×105–1×106 cells/mL. After centrifugation, the supernatant was discarded and cells were washed once with PBS. The collected cells were mixed with 1 mL DNA staining solution and 10 μL permeabilization solution by vortexing prior to 30 mins incubation at room temperature in the dark. The cells were sorted using a FACS Calibur machine (BD Biosciences, USA) and cell-cycle profiles were analyzed by ModFit 4.0 software. The experiments were repeated 3 times.
In vitro scratch assay
The cell suspensions were prepared and seeded in a 6-well plate at density of 1×105 cells per well and placed in a cell culture incubator overnight. The next day, a 10 μL pipette was used to scribe vertically in a 6-well plate well, the residue cell debris was washed three times with PBS, and then serum-free medium was added. The 6-well plate was placed in an incubator for continuous incubation, the scratched area was photographed at 0 and 24 hrs from the beginning when scratch was formed. The distance between the 2 edges of the gap space was measured for analysis. The experiments were repeated 3 times.
Transwell invasion assay
The Matrigel concentration was diluted to 1 mg/mL with serum-free medium, 24 μg of matrigel was uniformly coated in each Transwell chamber, and then allowed to solidify by placing at 37°C for 6 hrs. The ERRα-knockdown LUAD cells or control LUAD cells were prepared as a cell suspension using a serum-free medium at a density of 40–50×105/mL, and 100 μL of the cell suspension was added to the upper well of the chambers. Complete medium containing FBS was added to the lower chambers. Culture in a cell culture incubator. After 24 hrs, the cells in the chambers that did not migrate through the polycarbonate membrane were gently wiped with a cotton swab, and then the residual liquid in the chambers was aspirated. The invaded cells were fixed with methanol at room temperature for 30 mins, and stained with 0.1% crystal violet, and kept at room temperature for 20 mins. The invaded cells were then rinsed with water and dried naturally. Under the microscope, 5 randomly selected fields of view were photographed to calculate the invaded cells. The experiments were repeated 3 times.
Establishment of a mouse model with LUAD
A mouse model of LUAD was constructed using the protocol of the Cold Spring Harbor Laboratory.17 BALB/c mice aged 21–28 d were purchased and divided into 2 groups, model group and control group, each group contained 10 mice. The mice were weighed and labeled. The model group received intraperitoneal injection with urethane (1 mg/g); the control group was intraperitoneally injected with the same amount of PBS. At the 40th week of feeding, the mice were sacrificed, their lung tissues were resected to observe any visible adenocarcinoma nodules. The generated mouse lung nodules were embedded in paraffin. HE staining of paraffin tissue sections was performed to verify the existence of cancer cells, thus confirming the successful construction of the lung cancer model.
Western blot analysis
Total protein was extracted and quantified using BCA Protein Assay Reagent (Thermo Fisher Scientific) to determine the amount of loading. Corresponding 5×loading buffer and protein samples were mixed by vortex. The prepared protein samples were denatured by heating at 100°C for 5 mins, cooled and then frozen in a −20°C refrigerator. Thirty micrograms of protein from each sample were separated by 10% SDS-PAGE gel (Invitrogen). After electrophoresis, the separated proteins were transferred to polyvinylidene difluoride (PVDF) membranes and blocked with 5% skim milk for 1 hr. The membrane was then incubated overnight at 4°C with the following primary antibody: ERRα (1:2000, abcam ab76228) and β-actin (1:3000, Santa Cruz Biotechnology). After the incubation, the strips were washed 3 times with TBST. The protein underwent incubation for 1 hr at room temperature with secondary antibody (1:3000, Santa Cruz Biotechnology). After the incubation of the secondary antibody, the strips were washed 3 times with TBST before the protein bands were visualized with ECL solution (Thermo Fisher Scientific). Gray value of the blots was quantified by ImageJ software. All the experiments have been repeated 3 times.
Immunohistochemical staining
Immunohistochemistry experiments were performed using the specific HRP/DAB (ABC) detection IHC kit (abcam ab64264). The lung cancer tissue chip (Shanghai Core Super Biotechnology Co., Ltd.) was dewaxed and hydrated, then heated with citric acid for retrieval of antigen. After antigen recovery, the chip was washed 3 times with PBS. Endogenous peroxidase activity was blocked by dropwise addition of 3% H2O2, followed by incubated for 10 mins at room temperature, and washed 3 times with PBS. The 5% BSA was incubated for 1 hr at room temperature and then blocked. Antibody of ERRα (abcam, ab93173) was diluted (1:200) and incubated at 4°C overnight. The sections were rewarmed for 1 hr and washed 3 times with PBS, followed by incubation with biotin-labeled secondary antibody for 10 mins at room temperature and washed 4 times with PBS. Horseradish peroxidase-labeled streptomycin avidin working solution was added dropwise, incubated for 10 mins at room temperature, and washed 4 times with PBS. The color reaction was then carried out using DAB. Hematoxylin was used for counterstain. Images were captured under a microscope. PBS was used instead of the primary antibody as a blank control.
Brown-yellow particles appearing in the nucleus indicate positive ERRα expression. The IRS scoring standard was adopted. The product of staining intensity (SI) and percentage of positive cells (PP), ie, IRS=SI×PP. SI can be divided into 4 levels, 0 is no positive cells, 1 is weakly positive, 2 is moderately positive, and 3 is strongly positive. PP can be divided into 5 grades, 0 grades are negative, 1 grade ≤10%, 2 grades 11–50%, 3 grades 51–80%, and grade 4>80%. When the product of SI and PP is >3, the result is positive for immunoreactivity, 3<product ≤5 is moderately positive, and product >5 is strongly positive. Immunohistochemical sections were observed under a microscope and scored in conjunction with the IRS scoring criteria.
Statistical analysis
Statistical analysis of the data was performed using spss20.0 statistical software. All values are reported as x±s for three independent experiments unless otherwise stated. Data were analyzed by two-tailed unpaired Student’s t-test between the two groups.
The immunohistochemical results of lung cancer tissue microarray were compared by t-test or Fisher exact probability method. The Kaplan–Meier method was used for single factor survival analysis, the log-rank test was used to calculate the survival difference among different groups, and the COX model was used for multivariate survival analysis. The prognostic variables (P<0.1) found in the univariate analysis were selected into the equation. The limit of the covariate into the multivariate equation was 0.05, and the bound of the multivariate equation was 0.1. In all statistical analyses, P<0.05 was considered statistically significant.
Statement
All the experiments are in accordance with the principles of good laboratory practice standards GB T 22278-2008. The animal studies were subjected to prior review and approval from the Ethics Committee of Shenzhen Second People’s Hospital. (Shenzhen, Guangdong, China). All animal experiments were carried out in the animal care facility of the Animal Laboratory of Shenzhen Center for Disease Control and Prevention, which is accredited by the Guangdong Provincial Department of Science and Technology (Guangzhou, Guangdong, China) and in accord with relevant institutional and Chinese national guidelines. The human studies were subjected to prior review and approval by the Ethics Committee of the Shanghai Outdo Biotech Company Ltd (SOBC), which was established in 2003 by the Chinese National Engineering Center for Biochip at Shanghai, located in Zhangjiang Hi-Tech Park of Pudong, Shanghai. The SOBC Ethics Committee is accredited by National Development and Reform Commission (China), and the SOBC follows the relevant Chinese national guidelines and regulations. All human research was conducted in accord with the most recent iteration of the Declaration of Helsinki and written informed consent was obtained from all the patients/patients’ families.
Results
The expression level of ERRα was found to be associated with survival prognosis in patients with LUAD
The correlations between ERRα mRNA expression level and survival information of 720 patients with LUAD (n=720) and 524 patients with LSCC (n=524) were analyzed by KM plotter, respectively. The results showed that patients with LUAD with high expression of ERRα had a worse prognosis (HR=1.68, P<0.001) (Figure 1A), while no significant correlation was found between the expression level of ERRα and the survival time of patients with LSCC (P=0.78) (Figure 1B).
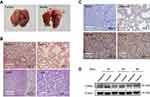
ERRα was upregulated in mouse model with LUAD
To further verify the expression of ERRα systematically, we established a mouse model with LUAD by intraperitoneal injection of Urethane. After the isolation of lungs, obvious tumor nodule could be observed in model mice while no formation of tumor nodule was found in control mice (Figure 2A). The results of the pathological analysis indicated tumor cell infiltration could be observed in model mice while no tumor cell infiltration could be found in control mice under the microscope after HE staining (Figure 2B). Immunohistochemistry analysis suggested that ERRα was in a higher level in cancerous tissues than in paracancerous tissues of model mice (Figure 2C). Moreover, analysis by Western blot detection also indicated an elevation of ERRα in model mice compared with control mice (P<0.05, Figure 2D).
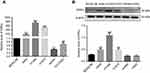
ERRα was found abnormally elevated in LUAD rather than LSCC
To evaluate ERRα in NSCLC, we analyzed the expressions of ERRα in normal lung bronchial epithelial cells (BEAS-2B), LUAD cells (A549, NCI-H1395, and NCI-H1975), and LSCC (SW900 and NCI-H520) cells by semi-quantitative PCR and Western blot, respectively. The results suggested that ERRα was upregulated in both A549, NCI-H1395, and NCI-H1975 cells while downregulated in SW900 and NCI-H520 cells compared with BEAS-2B (P<0.05, Figure 3).
Establishment of ERRα-knockdown LUAD cells
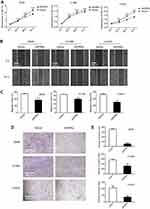
To further investigate the role of ERRα in the development of LUAD, we constructed ERRα-shRNA recombinant lentivirus plasmid. Then, we performed stable ERRα-knockdown on A549, NCI-H1395, and NCI-H1975 cells by lentivirus-mediated transfection, respectively. The expression levels of ERRα were verified by qPCR and Western blot analysis. Results suggested that ERRα was significantly suppressed through transfection of ERRα-shRNA in both mRNA and protein level (Figure 4).
ERRα promotes the proliferation, migration, and invasion of LUAD cells
To evaluate the role of ERRα in the development of LUAD, cell viabilities of ERRα-knockdown LUAD cells and corresponding LUAD cells transfected with control vector were measured. The results suggested that after knockdown of ERRα, the viability of LUAD cells was significantly suppressed (P<0.001, Figure 5A). As an important malignant indicator of LUAD, the effects of ERRα on cell migration were analyzed by scratch wound healing assay. The closure of scratch gaps in cells with ERRα-knockdown was significantly suppressed compared to control (P<0.001, Figure 5B and C). Moreover, we explored the impacts of ERRα on another key factor of malignancy, cell invasion through transwell invasion assay. The number of invaded cells was significantly reduced in ERRα-knockdown LUAD cells as compared to control (P<0.01, Figure 5D and E).
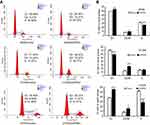
Knockdown of ERRα leads to cell cycle arrest at G2/M phase of LUAD cells
To further investigate the impacts of ERRα on cell cycle, fluorescence-activated cell sorting (FACS) assay was applied to analyze ERRα-related cell cycle distribution. As shown in Figure 6, the accumulation of ERRα-knockdown cells in the G2/M phase was higher compared with control (P<0.05, Figure 6) suggesting that knockdown of ERRα could lead to LUAD cells arresting in the G2/M phase.
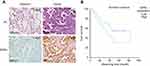
High positive rate of ERRα was found in clinical samples with LUAD
To investigate the role of ERRα in clinical outcomes, we analyzed the expression level of ERRα in clinical samples. Immunohistochemistry staining suggested higher expression of ERRα in cancerous tissues compared with paracancerous tissues (Figure 7A). ERRα is mainly located in the nucleus of tumor cells. The positive rates of ERRα in cancerous and paracancerous tissues were 78.2% (72/92) and 43.2% (38/88), respectively. The positive rate of ERRα in cancerous tissues was nearly 2-fold of which in paracancerous tissues (χ2=21.425, P<0.001).
High level of ERRα in tumor tissue was positively associated with poor prognosis
Based on the observation that higher positive rate of ERRα was found in cancerous tissues, we further evaluate the associations between ERRα and clinical outcomes of LUAD. The correlation between clinicopathological parameters and ERRα protein expression in individuals with LUAD is summarized in Tables 1 and 2. Both TNM stage and lymph node metastasis were found significantly correlated to ERRα (χ2=7.345, P=0.007; χ2=6.293, P=0.012). The expression of ERRα was higher in stage III–IV lung cancer than in stage I–II lung cancer. The difference was statistically significant. None of the following parameters such as age, gender, pathological grade, T stage, and distant metastasis were found correlated to ERRα (P>0.05, Table 1).
 |
Table 1 Relationship between ERRα expression and clinicopathological parameters in lung cancer |
 |
Table 2 Univariate analysis of prognosis in lung adenocarcinoma by Kaplan–Meier method |
Univariate analysis of Kaplan–Meier’s method showed (Table 2), with α=0.05 as the test level, and the prognostic-related variables were T-state (P=0.005), lymph node metastasis (P=0.036), TNM staging (P=0.046), ERRα classification (P=0.049), and ERRα high and low expression (P=0.041). Among them, the ERRα expression was divided into two groups: low expression (IRS score<6) and high expression (IRS score≥6). Survival analysis indicated that individuals with high expression of ERRα had lower prognosis (P=0.041), and survival curves were shown in Figure 7B. Furthermore, COX multivariate analysis indicated that when the prognostic factors (P<0.1) in the univariate analysis were included in the multivariate analysis, α=0.05 was used as the test level, only T stage (P=0.015) and ERRα expression (P=0.013, HR=1.597) were statistically significant (Table 3).
 |
Table 3 Multivariate analysis of prognosis in lung adenocarcinoma by COX model |
Discussion
ERRα is an important estrogen-related receptor which participates in key biological processes of multiple tumors. We found that ERRα was abnormally elevated in LUAD cells (A549, NCI-H1395, and NCI-H1975) rather than LSSC cells (SW900 and NCI-H520). These observations were further supported by the elevated ERRα expression in lung cancer mouse model. Given that the foregoing, we further knocked down ERRα in LUAD cells. Functional studies demonstrated that knockdown of ERRα impairs cell proliferation, migration, and metastasis of LUAD cells. Moreover, evaluation of ERRα in LUAD tissues indicated that high expression of ERRα positively associates with poor prognosis in LUAD patients, suggesting that ERRα is an independent risk factor for poor prognosis.
We confirmed abnormal elevation of ERRα in LUAD at both cellular, animal, and clinical levels. Our current results are consistent with another report about high expression of ERRα in LUAD cells.16 Although ERRα was reported to trigger proliferation and migration of NSCLC cells via interleukin-6,18 the exact role that ERRα plays on specific subtypes of lung cancer, such as adenocarcinoma and squamous cell carcinoma, has remained obscured. In our current study, we put forward that high expression of ERRα might only reside in LUAD. Moreover, for the prognosis analysis, the analysis results of public database (TCGA, EGA) that high mRNA level of ERRα was associated with poor prognosis in LAUD are consolidated in our current research at protein level by immunohistochemistry. The results of database analysis showed that high mRNA level of ERRα was a poor prognostic factor in LUAD, but there was no significant correlation between the high expression of ERRα and the prognosis of LSCC. It has been reported that mitochondrial regulator ERRα could be downregulated under the action of XCT-790, an estrogen-related receptor alpha inverse agonist, thus inducing production of mitochondrial reactive oxygen species and inhibiting the growth of lung cancer cells19 Further study indicated that ERRα has higher expression level in tissues with higher energy demand (such as heart, kidney, intestine, and brown fat), but lower expression levels in organs with lower energy demand, such as liver, lung, and vagina.20,21 Cell proliferation, migration, and invasion are energy-consuming processes, which might be responsible for ERRα-mediated malignant properties in LUAD cells. ERRα has been reported closely related to the occurrence and development of tumors.22 It has been known that ERRα could affect the cell cycle by participating in the estrogen signal transduction system and could regulate the transcriptional activity of target genes, which further regulates the quality and function of mitochondria,23,24 the biological behaviors such as proliferation, invasion, and migration of cancer cells were also affected accordingly.22,25 Furthermore, ERRα was found involved in the process of tumorigenesis, metastasis, and drug resistance of breast and prostate cancers.26,27 Interference with ERRα can exert anti-tumor effects by affecting multiple signaling pathways. Studies have found that ERRα-induced metabolic reprogramming can promote the survival of lapatinib-resistant cancer cells and demonstrates the potential of ERRα inhibition as adjunctive therapy for poor prognosis of HER2-positive breast cancer.26 High expression of ERRα was also found associating with poor prognosis in breast cancer,28 prostate cancer,29 colon cancer,30 ovarian cancer,31,32 endometrial cancer, and cervical cancer,33 while its association with LUAD has not yet been profiled. In this study, the positive expression of ERRα in lung cancer tissues was significantly higher than in paracancerous tissues. With the appearance of lymph node metastasis and the higher TNM stage and T stage, the expression of ERRα in lung cancer is higher, suggesting that ERRα may participate in and play an important role in tumor proliferation, invasion and metastasis, and the microenvironment of tumor changes. Patients with high expression of ERRα had poor prognosis (P=0.041), and high expression of ERRα was a prognostic risk factor (P=0.013, HR=1.597). The results indicate that the death risk of ERRα high expression is 1.597 times higher than ERRα low level in LUAD patients. This finding may provide a novel perspective for prognosis evaluation of LUAD patients. The prognosis of lung cancer is a key issue for clinicians. Although the related research has gradually increased in recent years, there is no uniform prognostic standard. The current prognostic indicators for evaluating lung cancer include epidermal growth factor receptor (EGFR), K-ras gene mutation, TTF-1, Napsin A34–36; however, these indicators certain limitations in specificity and sensitivity. ERRα may be able to perform an auxiliary assessment in cases where other indicators are not valid, especially in distinguishing between LUAD and LSCC, which can play an important role in clinical application.
In conclusion, ERRα was identified as an aggressive factor through mediating proliferation, migration, invasion, and cell cycle arrest at G2/M phase of LUAD cells. Further study suggested ERRα as an independent risk factor of poor prognosis in LUAD. Our findings indicate the key role of ERRα in LUAD as a potential target for prognosis, diagnosis, and treatment.
Acknowledgment
This work was supported by Natural Science Foundation of Guangdong (grant numbers 2016A030313029, 2017A030313668), Sanming Project of Medicine in Shenzhen (grant number SZSM201612031) and Shenzhen Municipal Government of China (grant numbers JCYJ20170817171808368, JCYJ20170818085657917, JCYJ20160328161613864, JSGG20170414104216477).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
2. Cheng I, Le GM, Noone AM, et al. Lung cancer incidence trends by histology type among Asian American, Native Hawaiian, and Pacific Islander populations in the United States, 1990–2010. Cancer Epidemiol Biomarkers Prev. 2014;23(11):2250–2265. doi:10.1158/1055-9965.EPI-14-0493
3. Travis WD, Brambilla E, Burke AP, Marx A, Nicholson AG. Introduction to the 2015 World Health Organization classification of tumors of the lung, pleura, thymus, and heart. J Thorac Oncol. 2015;10(9):1240–1242. doi:10.1097/JTO.0000000000000663
4. Giguere V. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr Rev. 2008;29(6):677–696. doi:10.1210/er.2008-0017
5. Giguere V, Yang N, Segui P, Evans RM. Identification of a new class of steroid hormone receptors. Nature. 1988;331(6151):91–94. doi:10.1038/331091a0
6. Deblois G, Giguere V. Oestrogen-related receptors in breast cancer: control of cellular metabolism and beyond. Nat Rev Cancer. 2013;13(1):27–36. doi:10.1038/nrc3396
7. Luo C, Balsa E, Thomas A, et al. ERRα maintains mitochondrial oxidative metabolism and constitutes an actionable target in PGC1α-elevated melanomas. Mol Cancer Res. 2017;15(10):1366–1375. doi:10.1158/1541-7786.MCR-17-0143
8. Ranhotra HS. The orphan estrogen-related receptor alpha and metabolic regulation: new frontiers. J Recept Signal Transduct Res. 2015;35(6):565–568. doi:10.3109/10799893.2015.1024853
9. Vanacker JM, Pettersson K, Gustafsson JA, Laudet V. Transcriptional targets shared by estrogen receptor- related receptors (ERRs) and estrogen receptor (ER) alpha, but not by ERβ. Embo J. 1999;18(15):4270–4279. doi:10.1093/emboj/18.15.4270
10. Charest-Marcotte A, Dufour CR, Wilson BJ, et al. The homeobox protein Prox1 is a negative modulator of ERR{α}/PGC-1{α} bioenergetic functions. Genes Dev. 2010;24(6):537–542. doi:10.1101/gad.1871610
11. Gallet M, Vanacker JM. ERR receptors as potential targets in osteoporosis. Trends Endocrinol Metab. 2010;21(10):637–641. doi:10.1016/j.tem.2010.06.008
12. Huss JM, Garbacz WG, Xie W. Constitutive activities of estrogen-related receptors: transcriptional regulation of metabolism by the ERR pathways in health and disease. Biochim Biophys Acta. 2015;1852(9):1912–1927. doi:10.1016/j.bbadis.2015.06.016
13. Casaburi I, Chimento A, De Luca A, et al. Cholesterol as an endogenous ERRα agonist: a new perspective to cancer treatment. Front Endocrinol (Lausanne). 2018;9:525. doi:10.3389/fendo.2018.00420
14. Tam IS, Giguere V. There and back again: the journey of the estrogen-related receptors in the cancer realm. J Steroid Biochem Mol Biol. 2016;157:13–19. doi:10.1016/j.jsbmb.2015.06.009
15. Xu Z, Wang Y, Xiao ZG, et al. Nuclear receptor ERRα and transcription factor ERG form a reciprocal loop in the regulation of TMPRSS2: eRGfusion gene in prostate cancer. Oncogene. 2018;37(48):6259–6274. doi:10.1038/s41388-018-0409-7
16. Huang JW, Guan BZ, Yin LH, et al. Effects of estrogen-related receptor alpha (ERRα) on proliferation and metastasis of human lung cancer A549 cells. J Huazhong Univ Sci Technolog Med Sci. 2014;34(6):875–881. doi:10.1007/s11596-014-1367-0
17. Gurley KE, Moser RD, Kemp CJ. Induction of lung tumors in mice with urethane. Cold Spring Harb Protoc. 2015;2015(9):pdbprot077446. doi:10.1101/pdb.prot077446
18. Zhang J, Guan X, Liang N, Li S. Estrogen-related receptor alpha triggers the proliferation and migration of human non-small cell lung cancer via interleukin-6. Cell Biochem Funct. 2018;36(5):255–262. doi:10.1002/cbf.3337
19. Wang J, Wang Y, Wong C. Oestrogen-related receptor alpha inverse agonist XCT-790 arrests A549 lung cancer cell population growth by inducing mitochondrial reactive oxygen species production. Cell Prolif. 2010;43(2):103–113. doi:10.1111/j.1365-2184.2009.00659.x
20. Bonnelye E, Vanacker JM, Dittmar T, et al. The ERR-1 orphan receptor is a transcriptional activator expressed during bone development. Mol Endocrinol. 1997;11(7):905–916. doi:10.1210/mend.11.7.9948
21. Heard DJ, Norby PL, Holloway J, Vissing H. Human ERRγ, a third member of the estrogen receptor-related receptor (ERR) subfamily of orphan nuclear receptors: tissue-specific isoforms are expressed during development and in the adult. Mol Endocrinol. 2000;14(3):382–392. doi:10.1210/mend.14.3.0431
22. Bianco S, Sailland J, Vanacker JM. ERRs and cancers: effects on metabolism and on proliferation and migration capacities. J Steroid Biochem Mol Biol. 2012;130(3–5):180–185. doi:10.1016/j.jsbmb.2011.03.014
23. Mootha VK, Bunkenborg J, Olsen JV, et al. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115(5):629–640. doi:10.1016/s0092-8674(03)00926-7
24. Soriano FX, Liesa M, Bach D, Chan DC, Palacin M, Zorzano A. Evidence for a mitochondrial regulatory pathway defined by peroxisome proliferator-activated receptor-gamma coactivator-1 alpha, estrogen-related receptor-alpha, and mitofusin 2. Diabetes. 2006;55(6):1783–1791. doi:10.2337/db05-0509
25. Ranhotra HS. Estrogen-related receptor alpha and mitochondria: tale of the titans. J Recept Signal Transduct Res. 2015;35(5):386–390. doi:10.3109/10799893.2014.959592
26. Deblois G, Smith HW, Tam IS, et al. ERRα mediates metabolic adaptations driving lapatinib resistance in breast cancer. Nat Commun. 2016;7:12156. doi:10.1038/ncomms12156
27. Zou C, Yu S, Xu Z, et al. ERRα augments HIF-1 signalling by directly interacting with HIF-1α in normoxic and hypoxic prostate cancer cells. J Pathol. 2014;233(1):61–73. doi:10.1002/path.4329
28. Suzuki T, Miki Y, Moriya T, et al. Estrogen-related receptor alpha in human breast carcinoma as a potent prognostic factor. Cancer Res. 2004;64(13):4670–4676. doi:10.1158/0008-5472.CAN-04-0250
29. Fujimura T, Takahashi S, Urano T, et al. Increased expression of estrogen-related receptor alpha (ERRα) is a negative prognostic predictor in human prostate cancer. Int J Cancer. 2007;120(11):2325–2330. doi:10.1002/ijc.22363
30. Liang R, Lin Y, Yuan CL, et al. High expression of estrogen-related receptor alpha is significantly associated with poor prognosis in patients with colorectal cancer. Oncol Lett. 2018;15(4):5933–5939. doi:10.3892/ol.2018.8011
31. Fujimoto J, Sato E. Clinical implication of estrogen-related receptor (ERR) expression in uterine endometrial cancers. J Steroid Biochem Mol Biol. 2009;116(1–2):71–75. doi:10.1016/j.jsbmb.2009.04.012
32. Sun P, Sehouli J, Denkert C, et al. Expression of estrogen receptor-related receptors, a subfamily of orphan nuclear receptors, as new tumor biomarkers in ovarian cancer cells. J Mol Med (Berl). 2005;83(6):457–467. doi:10.1007/s00109-005-0639-3
33. Mori T, Sawada M, Kuroboshi H, et al. Estrogen-related receptor α expression and function are associated with vascular endothelial growth factor in human cervical cancer. Int J Gynecol Cancer. 2011;21(4):609–615. doi:10.1097/IGC.0b013e3182017e9b
34. Rekhtman N, Pietanza CM, Sabari J, et al. Pulmonary large cell neuroendocrine carcinoma with adenocarcinoma-like features: napsin A expression and genomic alterations. Mod Pathol. 2018;31(1):111–121. doi:10.1038/modpathol.2017.110
35. Warth A, Penzel R, Lindenmaier H, et al. EGFR, KRAS, BRAF and ALK gene alterations in lung adenocarcinomas: patient outcome, interplay with morphology and immunophenotype. Eur Respir J. 2014;43(3):872–883. doi:10.1183/09031936.00018013
36. Yang M, Nonaka D. A study of immunohistochemical differential expression in pulmonary and mammary carcinomas. Mod Pathol. 2010;23(5):654–661. doi:10.1038/modpathol.2010.38
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.