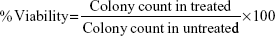Back to Journals » International Journal of Nanomedicine » Volume 11
Epigenetic modulation upon exposure of lung fibroblasts to TiO2 and ZnO nanoparticles: alterations in DNA methylation
Authors Patil N, Gade WN, Deobagkar DD
Received 12 April 2016
Accepted for publication 1 June 2016
Published 7 September 2016 Volume 2016:11 Pages 4509—4519
DOI https://doi.org/10.2147/IJN.S110390
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Thomas Webster
Nayana A Patil,1,2 WN Gade,2 Deepti D Deobagkar1
1Department of Zoology, Molecular Biology Research Laboratory, Centre of Advanced Studies, 2Department of Biotechnology, Proteomic Research Laboratory, Savitribai Phule Pune University, Pune, India
Abstract: Titanium dioxide (TiO2) and zinc oxide (ZnO) nanoparticles (NPs) are promising candidates for numerous applications in consumer products. This will lead to increased human exposure, thus posing a threat to human health. Both these types of NPs have been studied for their cell toxicity, immunotoxicity, and genotoxicity. However, effects of these NPs on epigenetic modulations have not been studied. Epigenetics is an important link in the genotype and phenotype modulation and misregulation can often lead to lifestyle diseases. In this study, we have evaluated the DNA methylation-based epigenetic changes upon exposure to various concentrations of NPs. The investigation was designed to evaluate global DNA methylation, estimating the corresponding methyltransferase activity and expression of Dnmt gene using lung fibroblast (MRC5) cell line as lungs are the primary route of entry and target of occupational exposure to TiO2 and ZnO NPs. Enzyme-linked immunosorbent assay-based immunochemical assay revealed dose-related decrease in global DNA methylation and DNA methyltransferase activity. We also found direct correlation between the concentration of NPs, global methylation levels, and expression levels of Dnmt1, 3A, and 3B genes upon exposure. This is the first study to investigate effect of exposure to TiO2 and ZnO on DNA methylation levels in MRC5 cells. Epigenetic processes are known to play an important role in reprogramming and adaptation ability of an organism and can have long-term consequences. We suggest that changes in DNA methylation can serve as good biomarkers for early exposure to NPs since they occur at concentrations well below the sublethal levels. Our results demonstrate a clear epigenetic alteration in response to metal oxide NPs and that this effect was dose-dependent.
Keywords: nanotoxicity, epigenetics, global DNA methylation, 5-mC, DNA methyltransferase, Dnmt
Introduction
Nanotechnology is growing at an exponential rate and undoubtedly is poised to change technology that offers multiple advantages. It is important to examine toxicological impact and consequences on health and environment when nanoparticles (NPs) are being used in numerous applications. A NP as compared with its bulky counterpart has novel/different physical properties such as an increased surface area to volume ratio, reactive sites, charge, shape, mobility, and thermal properties.1,2 Several processes have been designed based on these as well as photocatalytic, scattering, and homogeneous distribution properties for commercial applications of NPs in cosmetics, sunscreens, drug delivery, protective coatings making it easy to clean surfaces, in food for sensing biochemical parameters, and in textiles making them stain- and insect-resistant, and water repellent.3–7 Many new types of nanomaterials are being released in significant amounts with wide variety of applications in areas such as medicine, diagnostics, drug delivery,8 and biosensors.9,10 The increasing applications of NPs in various fields have led to an increase in human exposure which in turn may lead to toxicological effects and unexpected health and environmental hazards.11,12 Among the intentionally engineered NPs, metal oxide NPs are the most widely produced and used nanomaterials. It has been demonstrated that exposure to metals often may lead to toxicity, altered gene expression, changes in epigenetic marks, and metal-induced carcinogenesis.13,14
Accumulating evidence clearly shows that exposure to NPs can be toxic to biological system ranging from prokaryotes to higher eukaryotes including humans. Several in vivo and in vitro studies have shown that exposure to NPs leads to an inflammatory response, DNA damage, oxidative stress, lipid peroxidation, apoptosis, micronuclei formation, altered gene expression, genotoxicity, cytotoxicity, reproductive toxicity, immunotoxicity, and nongenotoxic carcinogenicity.15–23 Different mechanisms for nongenotoxic carcinogenicity have been proposed, one of these is the epigenetic changes in the DNA methylation pattern that may result in altered gene expression.24 In parallel, genomics25 and proteomics26 data have suggested altered gene and protein profile under NP-exposed scenario; however, epigenetic variation has gained relatively little attention.
Epigenetics involves stable and heritable changes in gene expression without changing DNA sequence. Epigenetic mechanisms principally include DNA methylation patterns, posttranslational modification of histones tails, chromatin remodeling, and microRNAs (miRNAs).27 Given that the epigenomic signatures can be propagated through cell division, epigenetic dysregulation may persist even after the exposure is removed, and if these changes remain undetected, it could lead to long-term deleterious effects in biological systems. For example, epigenetic dysregulation is central to the initiation and progression of some cancers. The global hypomethylation is seen in a number of cancers, including thyroid, breast, cervical, prostate, stomach, lung, bladder, esophagus, colorectum, and liver.28–30
Rapidly growing evidence has linked exposure to NP with epigenetic variations,31 including changes in DNA methylation, histone modifications, and miRNAs.32 Some of such epigenetic changes have been associated with variation in gene expression. For example, cadmium telluride quantum dots and silica NPs are reported to affect global DNA methylation pattern, modulate DNA methyltransferase activity, methyl-CpG-binding domain (MBD) protein expression, and/or alter posttranslational modifications of histone proteins.33–35 SiO2 NP has been shown to decrease the messenger RNA (mRNA) expression of PARP-1, increase the DNA methylation levels of PARP-1 promoter, decrease global DNA methylation, and the related methyltransferase, including Dnmt1, Dnmt3A, and MBD2.35,36 Similar study on silver NPs (AgNPs) shows that at sublethal levels AgNP can alter histone methylation, thereby effecting globin gene expression in red blood cells.37 Copper oxide and gold NPs are shown to induce alterations in miRNA expression.38–40 Recent study has reported that short-term exposure to engineered NPs leads to epigenetic alterations and an increase in L1 and Alu/SINEs mRNA transcripts in macrophages and lung epithelium.41 It has also been demonstrated that workplace exposure to NPs and their associated volatile chemicals can induce global demethylation, especially of retrotransposons in LINE and SINE sequences. NPs can lead to increase in reactive oxygen species production and oxidative DNA damage, which may affect the ability of methyltransferases activity leading to DNA hypomethylation and altered expression of methylation-regulated genes.42 However, there are no reports on the influence of titanium dioxide (TiO2) and zinc oxide (ZnO) NP on epigenetic integrity at sublethal concentration. TiO2 and ZnO NPs are considered as photocatalysts, and are extensively used in cosmetics and sunscreens.43 TiO2 and ZnO NPs are also used in paints, papers, toothpastes, food products, outdoor furniture varnishes, surface coating, textiles, and plastics.44,45
In the present study, we have examined the effect of sublethal concentration of TiO2 and ZnO NPs on modulation of global DNA methylation and dynamic alteration of DNA methyltransferases. The occupational exposure of both TiO2 and ZnO NPs is known to mainly affect lungs, therefore, lung fibroblast (MRC5) cell line was used as a model to determine the potential modulations in DNA methylation. Here, we report that sublethal concentration of TiO2 and ZnO NPs can induce epigenetic changes, which may lead to reprogramming of broad spectrum of gene expression.
Materials and methods
Chemicals
TiO2 (634662) and ZnO (544906) NPs were purchased from Sigma-Aldrich (Pune, India) and used for the experiments. Dulbecco’s Modified Eagle’s Medium (DMEM) and 0.25% trypsin–ethylenediaminetetraacetic acid were purchased from Invitrogen (Carlsbad, CA, USA). Fetal bovine serum was purchased from Life Technologies (Waltham, MA, USA). Penicillin–streptomycin was purchased from Life Technologies. The (4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, M5655) was purchased from Sigma-Aldrich (India).
Cell culture and exposure to NPs
Lung fibroblast (MRC5) cells were provided by American Type Culture Collection (ATCC, Manassas, VA, USA). The cell line (MRC5) was cultured in DMEM supplemented with 10% fetal bovine serum and 100 U/mL penicillin–streptomycin at 37°C and 5% CO2. NPs were suspended in culture medium at a concentration of 1 mg/mL, and then sonicated for 5 minutes. The solution was then diluted with medium to a concentration of 10 μg/mL. The dilutions of NPs were vigorously vortexed for 30 seconds prior to cell exposure to avoid NP agglomeration. Cells were grown to 80% confluency, monolayer cells were trypsinized by using 0.25% trypsin–ethylenediaminetetraacetic acid solution and seeded in 96- or 24-well plates (Corning Incorporated, Corning, NY, USA) at a density of 1×104 cells/well or 5×104 cells/well, and allowed to attach for 12 hours. The NPs were then diluted to appropriate concentrations and immediately applied to the cells. Dose-dependent cytotoxicity was assessed by exposing cells to the TiO2 and ZnO NPs at concentrations of 0, 0.125, 0.25, 0.5, 1, 2, 4, and 8 μg/mL for 24, 48, and 72 hours.
MTT assay
Viability of the cells exposed to metal oxide NPs for three different time points ranging from 24 to 72 hours was evaluated using the MTT reduction method.46 After incubation, the cell culture media was aspirated; 150 μL of MTT (5 mg/mL) was added to each well and incubated for 4 hours, 200 μL dimethyl sulfoxide was then added to dissolve the dark blue crystal. An optical density of 492 nm was used to monitor cell viability.
Colony-forming assay
The lung fibroblast cells (50 cells/well) were seeded into a 24-well plate and incubated overnight. These cells were treated with different concentration (0–8 μg/mL) of NPs for 10 days, medium was removed, and cells were fixed using chilled ethanol. Cells were stained with crystal violet (0.1% in ethanol) for 30 minutes. The plate was washed with water and allowed to dry. Numbers of colonies were counted. Percent viability was calculated using the following formula:
|
Total glutathione assay
The change in glutathione (GSH) level of NP-exposed cells was determined using a commercially available tGSH kit (Sigma-Aldrich) following manufacturer’s instruction. Briefly, NP-treated cells were sonicated in 5% sulfosalicylic acid. The samples were centrifuged at 10,000 rpm for 15 minutes at 4°C, and the supernatant was collected. The cell extract (20 μL) was transferred to a 96-well plate, and 150 μL of freshly prepared assay cocktail was added. The plates were then incubated for 5 minutes, then 50 μL of nicotinamide adenine dinucleotide phosphate solution (0.16 mg/mL) was added and the absorbance was read at 412 nm using a microplate reader (Multiskan EX; Thermo Scientific, Waltham, MA, USA). Standard curve was prepared using 50 μL of standards having total glutathione (tGSH) equivalents ranging from 0 to 50 μM. tGSH for each sample was then calculated from the standard curve as nmol/mg protein.
Nucleic acid isolation and quantitative real-time polymerase chain reaction
Total genomic DNA was isolated from NP-treated and NP-untreated cells at 24 and 48 hours exposure by sodium dodecyl sulfate/proteinase K digestion and phenol-chloroform extraction method. RNA was isolated from 24 hours NP-treated and NP-untreated cells using High Pure RNA Isolation Kit (Roche, Mannheim, Germany) and spectrophotometrically assessed for its quality. To remove chromosomal DNA, RNA was treated with DNase (Ambion, Oberursel, Germany). The complementary DNA (cDNA) was synthesized from RNA (1 μg) using Verso cDNA synthesis kit (Thermo Scientific). In brief, approximately 150–200 bp sequence from the Dnmt genes was amplified using specific primer sets (Table 1). The polymerase chain reaction (PCR) (20 μL) containing cDNA template (1 μL) in KAPA SYBR FAST qPCR Master Mix (2X) (Kapa Biosystems, Salt River, Cape Town, South Africa) with gene-specific primers (3 pM) was performed using ABI StepOne real-time PCR machine (Applied Biosystems, Waltham, MA, USA). The program details were 95°C, 2 minutes, followed by 94°C, 15 seconds → 60°C, 15 seconds → 72°C, 20 seconds (40 cycles). All qPCR reactions were performed in triplicate, and the mean of the three reactions was considered as a representative value for each sample. The expression levels of Dnmt1, 3A, and 3B were normalized to the endogenous control, 18S rRNA gene whose intensity did not change in treated cells as compared with untreated cells. Melting curve analysis was performed at the end of amplification. Comparative CT method was used to calculate relative expression of individual genes.
  | Table 1 List of primers used in this study |
Immunochemical staining for the detection of 5-methylcytosine
Genomic DNA was isolated from NP-treated and NP-untreated MRC5 cell line by phenol chloroform method. The method as described47,48 was used for detection of 5-methylcytosine (5-mC). MRC5 genomic DNA (500 ng) was spotted on nitrocellulose membrane (Schleicher & Schuell), dried and cross-linked by exposure to ultraviolet on both sides of the membrane for 3 minutes. Anti-5-mC antibody49 was used at a dilution of 1:1,000. The antibody reaction was visualized by using peroxidase conjugate of antirabbit antibody followed by staining for peroxidase activity using diaminobenzidine tetrahydrochloride (Sigma-Aldrich Co., St Louis, MO, USA) and H2O2 (Merck, Darmstadt, Germany).
Quantification of the 5-mC content in genomic DNA
The genomic DNA cytosine methylation level in cell line exposed to NPs at 24 and 48 hours was assessed by using an enzyme-linked immunosorbent assay-based commercial kit (MDQ1, Imprint® Methylated DNA Quantification Kit, Sigma-Aldrich). DNA at a concentration of 150 ng was diluted with 30 μL of binding buffers and incubated at 60°C. The samples were incubated with capture and detection antibodies and absorbance was read at 450 nm. Quantification of DNA methylation was obtained by calculating the amount of methylated cytosines in the sample relative to methylation in a positive control, which was provided by the manufacturer.
DNA methyltransferase activity
For the estimation of methyltransferase activity, cells were collected at 24 and 48 hours incubation with NPs, pelleted (5500 rpm, 5 minutes), and lysed in a phosphate-buffered saline by sonication. Cells were sonicated using the following setting: 70 Hz, 0.7 seconds cycle, five repetitions for 1 minute. The sonicated cells were then centrifuged at 14,000 rpm for 30 minutes at 4°C. The supernatant was collected and protein concentrations were determined in the cell lysates with a Bradford reagent (Bio-Rad, Laboratories Inc., Hercules, CA, USA) using bovine serum albumin as the standard. Cell lysates were used for determination of DNA methyltransferase activity using the EpiSeeker Dnmt Activity Quantification Assay Kit (Abcam, Cambridge, UK). In this assay, 5 μg of cell extract was incubated with a universal Dnmt substrate coated onto microplate wells. The methylated DNA was recognized with anti-5-mC antibody and then probed with detection antibodies provided with the kit. The amount of methylated DNA, which is proportional to enzyme activity, was then colorimetrically quantified using an enzyme-linked immunosorbent assay like reaction. DNA methyltransferase activity analysis was repeated thrice on each sample to minimize the assay variability. The results were expressed as absorbance units OD450 nm/h/mg protein.
Statistical analysis
All experiments were done at least three times unless otherwise indicated. Data are expressed as mean ± standard error, and statistical significance was tested among and between groups using one-way analysis of variance. Differences with P<0.05 were considered to be statistically significant.
Results
Cytotoxicity induced by the TiO2 and ZnO NPs
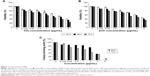
To evaluate the possible toxicity of TiO2 and ZnO NPs on cells, cell viability was determined after exposing MRC5 cells to TiO2 and ZnO NPs (0, 0.125, 0.25, 0.5, 1, 2, 4, and 8 μg/mL) for 24–72 hours. As shown in Figure 1, with the increasing dosage and time of incubation, viability of MRC5 cells was lower than control. At all time point of exposure to NPs, concentration ranging from 0.125 to 8 μg/mL TiO2 NP was more toxic as compared with ZnO NPs, significantly reducing the viability of MRC5 cells (Figure 1A and B). As a result, sublethal concentration with 80% viability and <2 μg/mL at 24 hours was chosen in current study, since this concentration and exposure would not induce high cytotoxicity to mask subtle epigenetic changes. The concentration at which cell viability is 80% is hereafter referred to as sublethal concentration, while 50% viability is hereafter referred to as lethal concentration.
Clonogenic survival
Clonogenic assay indicates the proliferative capacity of a single cell upon NP exposure for 10 days. The results of the colony-forming assay for cells treated with the TiO2 and ZnO are presented in Figure 1C. It represents the dose–response indicating the decrease in the colony number with the increasing concentration of both the NPs, thus demonstrating the effective inhibition of growth and proliferation of the cell line.
Oxidative stress
GSH status is considered to be an important oxidative stress marker in most cells. As shown in Figure 2, no significant reduction in tGSH level was observed even after exposure to 8 μg/mL of TiO2 and ZnO for 24 hours, indicating no obvious adaptive cell response of MRC5 with both NPs. Thus, both NPs did not trigger statistically significant difference in oxidative stress up to 8 μg/mL. Intracellular tGSH was reduced from 0.83 nmol/mg protein to 0.46 and 0.49 nmol/mg protein with 8 μg/mL concentration for TiO2 and ZnO NPs, respectively (Figure 2).
Analysis of global DNA methylation
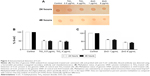
Global DNA methylation can be addressed by measuring the levels of 5-mC in control and exposed cells using an immunochemical detection method. Hypomethylation of genomic DNA was seen in lung fibroblast cells after exposure to both NPs (Figure 3A). When cells were treated with sublethal (0.5 μg/mL) and lethal concentration (4 μg/mL) of TiO2 NP, the DNA methylation was reduced to 49% and 46%, respectively, in comparison to control at 24 hours as estimated using the Imprint® Methylated DNA Quantification Kit (Figure 3B). At 48 hours, there was slight decrease in DNA methylation which corresponds to 48% and 45% for 0.5 and 4 μg/mL respectively. Similarly genomic DNA methylation was lower in the ZnO-exposed cells compared with the control. The average percent of methylated cytosine in DNA methylation was decreased to 59% and 58.8% at 24 and 48 hours incubation with 1 μg/mL (sublethal concentration) as compared with control. Interestingly, modest hypomethylation to 43% and 42% was also identified in genomic DNA after 24 and 48 hours exposure of cells to 8 μg/mL concentration of ZnO NPs (Figure 3C). Thus, it can be hypothesized that exposure to various concentrations of NP may differentially affect the methylation status of the genomic DNA, thereby giving differential cell response and toxicity levels at different concentration, but there is no significant difference with respect to time of incubation. These results suggest that NP leads to global DNA hypomethylation immediately after 24 hours exposure and subsequent exposure over the time does not result in further decrease in methylation.
Effects of NP exposure on DNA methyltransferase activity
Taking into account the observed alterations in genomic DNA methylation, to examine the effect of exposure of cells to NP on DNA methyltransferase activity, enzyme assay was carried out following exposure to TiO2 and ZnO NP for 24 and 48 hours. MRC5 cells were exposed to sublethal concentration 0.5 and 1 μg/mL of TiO2 and ZnO, respectively for 24 and 48 hours and DNA methyltransferase activity was determined. TiO2 and ZnO at these concentrations were not toxic to the cells. But at these concentrations, DNA methyltransferase activity was significantly repressed by almost 50% and 40% for TiO2 and ZnO, respectively, at 24 hours (Figure 4A and B). At lethal concentration (50% viability), that is, at 4 μg/mL concentration of TiO2 NP the repression of DNA methyltransferase activity was more pronounced, which led to 64% decrease as compared with control, while at 8 μg/mL concentration of ZnO NP the repression of DNA methyltransferase activity was decreased to 48% as compared with control (Figure 4A and B). Subsequent exposure for 48 hours does not result in further decrease in DNA methyltransferase activity. It is interesting to note that along with the decrease seen in level of genomic DNA methylation in NP-exposed cells, DNA methyltransferase activity was hampered in a concentration-dependent fashion.
NP treatment decreases Dnmt mRNA levels
To establish whether the observed changes in DNA methylation were accompanied by persistent changes in expression of the known Dnmt genes, mRNA levels of maintenance and de novo methyltransferase from MRC5 cells treated with TiO2 or ZnO NP for 24 hours were analyzed. Exposure to sublethal and lethal concentration of TiO2 and ZnO NP resulted in a marked reduction of the endogenous mRNA levels of Dnmt1, 3A, and 3B in MRC5 cell lines (Figure 5). The observed fold change reduction at 24 hours for Dnmt1 was 0.45–0.5 and 0.63–0.69 in TiO2- and ZnO-treated cells, respectively (Figure 5A); for Dnmt3B, relative expression was 0.49–0.5 in TiO2-treated cells and 0.61–0.68 in ZnO-treated cells (Figure 5B); for Dnmt3A, relative expression was 0.75–0.78 and 0.98 in TiO2- and ZnO-treated cells, respectively, compared with their respective controls (Figure 5C). Consistent with genomic DNA methylation and methyltransferase activity results, a significant downregulation of endogenous Dnmt1, 3A, and 3B expression levels was observed in cell exposed to both the NPs.
Discussion
Under conditions of occupational exposure, inhalation of TiO2 NPs is normally the principal route for entry into the human body.50 In terms of risk for human on exposure to TiO2 and ZnO NPs, lungs are certainly one of the major organs that are affected by occupational exposure. Thus, the study of lung fibroblast cell upon NP exposure is of great significance to understand human exposure situation. Therefore, we have chosen MRC5 cells as a model system to study the effect of TiO2 and ZnO NP on genomic DNA hypomethylation.
Currently, several in vitro approaches have linked engineered NPs with oxidative stress production, activation of inflammatory pathways, cytotoxicity, genotoxicity, immunotoxicity, gene expression modulation, and proteomic variation.15,19–23,29,30 However, the effect of NP on epigenetics/DNA methylation is not been fully investigated. The health effects associated with human exposure to NPs remain elusive and very little has been done with respect to investigating the epigenetic effects of nanomaterials. In the present investigation, we report that exposure to TiO2 and ZnO NP at concentrations that lead to nominal cytotoxicity (20% lethality) (Figure 1) can have adverse epigenetic effects at sublethal concentration. Very low-level exposures to these NPs can cause epigenetic changes.
Notably, NPs are known to stimulate oxidative stress, which in turn is associated with gene hypomethylation. Numerous studies have identified tGSH measurement as an indicator of oxidative stress.51 Thus to measure the levels of oxidative stress, tGSH content was measured after NP exposure in the range of 0.125–8 μg/mL concentration of TiO2 and ZnO NP (Figure 2). To identify the possible contribution of oxidative stress in DNA hypomethylation, 0.5 and 4 μg/mL concentration of TiO2 and 1 and 8 μg/mL concentration of ZnO were selected for further studies.
In our study, we observed decrease in genomic DNA methylation (Figure 3) along with corresponding DNA methyltransferase activity (Figure 4) at the above-mentioned concentration of TiO2 and ZnO NP. To the best of our knowledge, this is the first study to link altered DNA methylation patterns upon exposure to TiO2 and ZnO NPs, which have wide range of applications. A dose-related decrease in methylation in comparison with the control sample was detected. This can range from a net hypomethylation of 45%–50% in cells exposed to TiO2 NP (Figure 3B) and 40%–60% in cells exposed to ZnO NP (Figure 3C). These results suggest that both NPs induced overall DNA hypomethylation in MRC5 cells and that the effect was dose-dependent.
We have compared the DNA methyltransferase enzyme activity in treated and control conditions. Our study was based on quantitative analysis of DNA methyltransferase activity using immunochemical methodology, which is highly reproducible and accurate at measuring small changes in DNA methylation. DNA methyltransferase activity was reduced by 40%–50% in TiO2 and ZnO NP-exposed cells (Figure 4). NP-exposed cells definitely lead to hypomethylation of genomic DNA; however, upon subsequent exposure, there was no significant change in percent of DNA hypomethylation and corresponding methyltransferase activity.
We investigated whether changes in expression of Dnmts could account for the observed changes in DNA methylation and correlated with alterations in DNA methyltransferase activity. We further examined the expression of methyltransferases at two different concentrations. RT-PCR analysis of mRNA expression levels was employed to identify the effects exerted by NP on DNA methylation machinery. The 24-hour exposure to NP resulted in substantial and dose-dependent decreases in mRNA levels of all three DNA methyltransferases – Dnmt1, 3A, and 3B (Figure 5). Subtle downregulation of DNA methyltransferases was also observed in response to sublethal and lethal concentration. The mRNA expression of Dnmt1 changed in a trend similar to that of Dnmt3B after exposure to the NP. Effects of NP exposure on Dnmt3A were more pronounced as compared with Dnmt1 and Dnmt3B. Expression of Dnmts was decreased at both 0.5 and 4 μg/mL for TiO2 and 1 and 8 μg/mL for ZnO at 24 hours. Treatment with NP was found to reduce not only the amount of genomic DNA cytosine methylation and activity of DNA methyltransferase, but also the amount of DNA methyltransferase transcripts. Thus, the gradual loss of methylation of MRC5 cell genome during NP treatment was at least partly attributable to a decrease in DNA methyltransferase activity. Thus, the decreased methylation in the genomic DNA was due to persistent repression of DNA methyltransferases.
Epigenetic studies after exposure to NP-containing merchandise and knowledge on its effect is still limited. Epigenetic changes induced by NPs might be the earliest events during nanotoxicity and may provide a plausible explanation and link between molecular mechanism and nanotoxicity. However, it is clear that there is a lack of research in the “nanoepigenetics” field. Limited data suggest that exposure to some NPs, such as cadmium telluride quantum dots, SiO2, AgNP, gold NP, multiwalled carbon nanotubes, copper oxide, and engineered NPs adversely affect epigenetic landscape.31–41 To our knowledge, global DNA methylation and Dnmt expression levels have not previously been investigated for TiO2 and ZnO NP. Our results suggest that global hypomethylation is associated with concentration of NPs. The findings from this study are intriguing and merit further investigation.
Epigenetic modifications are extensively studied and used as potential biomarkers in several disease conditions.52,53 Most epigenetic biomarkers discovered to date are based on DNA methylation leading to molecular alteration in many human diseases. Numerous evidences show that methylated DNAs or miRNAs can serve as biomarkers for the early detection in tumorigenesis and many diseases such as diabetes, obesity, neurological, cardiovascular, genetic, psychiatric disorders, and stress.54–57
Our understanding of nanoepigenetics is increasing, but it is still far from complete, a gene-specific epigenomic and promoter DNA methylation pattern deserves further study. The resulting information might lead to the identification of potential NP exposure biomarkers for early detection of nanotoxicity.
Nanoepigenetics and exposure biomarkers integrated with genomic and/or proteomic studies can be a rich source of information that can ultimately result in a much improved understanding of molecular mechanisms for nanotoxicity. It also suggests that DNA hypomethylation can be used as an early exposure warning sign as a biomarker.
Conclusion
Significant correlation was noticed between NP exposure, endogenous DNA methyltransferase transcript levels, and global genomic DNA methylation along with activities of DNA methyltransferase enzyme. The repressed Dnmts with maintenance or de novo methyltransferase activity contribute to global hypomethylation. These data provide firm evidence that NPs even at sublethal concentrations at which no obvious cytotoxic effect or oxidative stress is manifested could still impede genomic DNA hypomethylation.
Aberrant global DNA hypomethylation is generally associated with chromosomal instability, overexpression of genes that is important in carcinogenesis, and modulation of expression of genes that would normally be tightly regulated by methylation.58 TiO2 has been classified as possible carcinogen to humans (group 2B) as well as TiO2 NPs induce tumorigenesis in animal models.59 Limited data shows that reactive oxygen species generation, inflammation, DNA damage, and signal alterations of certain cancer-related genes may be involved in the carcinogenicity of TiO2 NPs. We hypothesized that TiO2 NPs may operate at the epigenetic level by influencing chromosomal stability and gene expression and may play an important role in the etiology of their carcinogenesis.
Large body of data links NP exposure to induce alterations in gene expression, altered protein profile and toxicity, irrespective of the model system used. However, the mechanisms by which NPs can lead to these toxic effect and adverse outcomes are not well characterized. Therefore, we like to propose that DNA hypomethylation can be associated with and could explain molecular mechanism of NP toxicity and carcinogenicity for TiO2 NPs. Currently, the toxicity of engineered NPs is assessed with a number of approaches. Nanotoxicity is studied with an array of in vitro methods such as genomics, transcriptomics, and proteomics approach; parallel epigenomic studies will provide a valuable platform to predict the toxic potential of nanomaterials. Based on the results of our investigation, we believe that the inclusion of an assessment of methylation status, with an emphasis on dose–response relationships, as a component of initial toxicity testing can help in better understanding of possible mechanisms underlying toxicity.
The mechanism by which NPs interfere with DNA methylation remains unclear. It is likely that hypomethylation regulates gene expression, thus future work aims to explore specific genes effected by NP-induced hypomethylation. Although the relation between nanotoxicity and DNA hypomethylation is unknown, the study adds TiO2 and ZnO to a growing list of NPs, which could cause epigenetic changes.
Acknowledgments
This research was funded by the UGC Dr DS Kothari Postdoctoral fellowship. WNG and DDD acknowledge the departmental (Biotechnology and Zoology) research grant and UPE Phase II Biotherapeutics grant.
Author contributions
NAP carried out all experiments and analyzed the data sets. WNG and DDD conceived the experiments, supervised, and provided major intellectual inputs. NAP, WNG, and DDD contributed in revising the article critically for important intellectual content. All authors have approved the final article. All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Li X, Liu W, Sun L, et al. Effects of physicochemical properties of nanomaterials on their toxicity. J Biomed Mater Res A. 2015;103:2499–2507. | ||
Gatoo MA, Naseem S, Arfat MY, Dar AM, Qasim K, Zubair S. Physicochemical properties of nanomaterials: implication in associated toxic manifestations. Biomed Res Int. Epub 2014 Aug 6. | ||
Wiechers JW, Musee N. Engineered inorganic nanoparticles and cosmetics: facts, issues, knowledge gaps and challenges. J Biomed Nanotechnol. 2010;6(5):408–431. | ||
Hofmann-Amtenbrink M, Grainger DW, Hofmann H. Nanoparticles in medicine: current challenges facing inorganic nanoparticle toxicity assessments and standardizations. Nanomedicine. 2015;11(7):1689–1694. | ||
Xue CH, Chen J, Yin W, Jia ST, Jian-Zhong MA. Superhydrophobic conductive textiles with antibacterial property by coating fibers with silver nanoparticles. Appl Surf Sci. 2012;258:2468–2472. | ||
Sawhney AS, Condon B, Singh KV, Pang SS, Hui GD. Modern applications of nanotechnology in textiles. Text Res J. 2008;78:731–739. | ||
Scott N, Chen H. Nanoscale science and engineering for agriculture and food systems. Ind Biotechnol. 2013;9(1):17–18. | ||
Parveen S, Misra R, Sahoo SK. Nanoparticles: a boon to drug delivery, therapeutics, diagnostics and imaging. Nanomedicine. 2012;8(2):147–166. | ||
Kalele SA, Kundu AA, Gosavi SW, Deobagkar DN, Deobagkar DD, Kulkarni SK. Rapid detection of Escherichia coli by using antibody-conjugated silver nanoshells. Small. 2006;2(3):335–338. | ||
Kalele SA, Ashtaputre SS, Hebalkar NY, et al. Optical detection of antibody using silica–silver core–shell particles. Chem Phys Lett. 2005;404(1–3):136–141. | ||
Srivastava V, Gusain D, Sharma YC. Critical review on the toxicity of some widely used engineered nanoparticles. Ind Eng Chem Res. 2015;54(24):6209–6233. | ||
Peralta-Videa JR, Zhao L, Lopez-Moreno ML, de la Rosa G, Hong J, Gardea-Torresdey JL. Nanomaterials and the environment: a review for the biennium 2008–2010. J Hazard Mater. 2011;186(1):1–15. | ||
Baccarelli A, Bollati V. Epigenetics and environmental chemicals. Curr Opin Pediatr. 2009;21:243–251. | ||
Zamudio RM, Ha HC. Environmental epigenetics in metal exposure. Epigenetics. 2011;6(7):820–827. | ||
Lewinski N, Colvin V, Drezek R. Cytotoxicity of nanoparticles. Small. 2008;4(1):26–49. | ||
Huang YW, Wu CH, Aronstam RS. Toxicity of transition metal oxide nanoparticles: recent insights from in vitro studies. Materials. 2010;3(10):4842–4859. | ||
Djuris AD, Leung YH, Ng MC, et al. Toxicity of metal oxide nanoparticles: mechanisms, characterization, and avoiding experimental artefacts. Small. 2015;11(1):26–44. | ||
Luque-Garcia JL, Sanchez-Díaz R, Lopez-Heras I, Camara C, Martin P. Bioanalytical strategies for in-vitro and in-vivo evaluation of the toxicity induced by metallic nanoparticles. TrAC Trends Anal Chem. 2013;43:254–268. | ||
Khanna P, Ong C, Bay BH, Baeg GH. Nanotoxicity: an interplay of oxidative stress, inflammation and cell death. Nanomaterials. 2015;5(3):1163–1180. | ||
Sarkar A, Ghosh M, Sil PC. Nanotoxicity: oxidative stress mediated toxicity of metal and metal oxide nanoparticles. J Nanosci Nanotechnol. 2014;14(1):730–743. | ||
Singh N, Manshian B, Jenkins GS, et al. Nanogenotoxicology: the DNA damaging potential of engineered nanomaterials. Biomaterials. 2009;30(23–24):3891–3914. | ||
Zolnik BS, González-Fernández Á, Sadrieh N, Dobrovolskaia MA. Nanoparticles and the immune system. Endocrinology. 2010;151(2):458–465. | ||
Lozano-Fernández T, Ballester-Antxordoki L, PérezTemprano N, et al. Potential impact of metal oxide nanoparticles on the immune system: the role of integrins, L-selectin and the chemokine receptor CXCR4. Nanomedicine. 2014;10(6):1301–1310. | ||
Herceg Z, Lambert MP, Veldhoven KV, et al. Towards incorporating epigenetic mechanisms into carcinogen identification and evaluation. Carcinogenesis. 2013;34(9):1955–1967. | ||
Singha N, Manshiana B, Gareth JS, et al. NanoGenotoxicology: the DNA damaging potential of engineered nanomaterials. Biomaterials. 2009;30(23–24):3891–3914. | ||
Matysiak M, Kapka-Skrzypczak L, Brzóska K, Gutleb AC, Kruszewsk M. Proteomic approach to nanotoxicity. J Proteomics. 2016;137:35–44. | ||
Aaron D, Goldberg C, Allis D, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128(4):635–638. | ||
Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31(1):27–36. | ||
Hardy TM, Tollefsbol TO. Epigenetic diet: impact on the epigenome and cancer. Epigenomics. 2011;3(4):503–518. | ||
Costa PM, Fadeel B. Emerging systems biology approaches in nanotoxicology: towards a mechanism-based understanding of nanomaterial hazard and risk. Toxicol Appl Pharmacol. 2016;299:101–111. | ||
Stoccoro A, Karlsson HL, Coppede F, Migliore L. Epigenetic effects of nano-sized materials. Toxicology. 2013;313(1):3–14. | ||
Mytych J, Wnuk M. Nanoparticle technology as a double-edged sword: cytotoxic, genotoxic and epigenetic effects on living cells. J Biomater Nanobiotechnol. 2013;4(1):53–63. | ||
Choi AO, Brown SE, Szyf M, Maysinger D. Quantum dot-induced epigenetic and genotoxic changes in human breast cancer cells. J Mol Med (Berl). 2008;86(3):291–302. | ||
Conroy J, Byrne SJ, Gun’ko YK, et al. CdTe nanoparticles display tropism to core histones and histone-rich cell organelles. Small. 2008;4(11):2006–2015. | ||
Gong C, Tao G, Yang L, Liu J, Liu Q, Zhuang Z. SiO(2) nanoparticles induce global genomic hypomethylation in HaCaT cells. Biochem Biophys Res Commun. 2010;397(3):397–400. | ||
Gong C, Tao G, Yang L, et al. Methylation of PARP-1 promoter involved in the regulation of nano-SiO2-induced decrease of PARP-1 mRNA expression. Toxicol Lett. 2012;209(3):264–269. | ||
Qian Y, Zhang J, Hu Q, et al. Silver nanoparticle-induced hemoglobin decrease involves alteration of histone 3 methylation status. Biomaterials. 2015;70:12–22. | ||
Balansky R, Longobardi M, Ganchev G, et al. Transplacental clastogenic and epigenetic effects of gold nanoparticles in mice. Mutat Res. 2013;42:751–752. | ||
Ng CT, Dheen ST, Yip WG, Ong CN, Bay BH, Yung LL. The induction of epigenetic regulation of PROS1 gene in lung fibroblasts by gold nanoparticles and implications for potential lung injury. Biomaterials. 2011;32(30):7609–7615. | ||
Yao Y, Costa M. Genetic and epigenetic effects of nanoparticles. J Mol Genet Med. 2013;7:86. | ||
Lu X, Miousse IR, Pirela SV, Melnyk S, Koturbash I, Demokritou P. Short-term exposure to engineered nanomaterials affects cellular epigenome. Nanotoxicology. 2016;10(2):140–150. | ||
Ziech D, Franco R, Pappa A, Panayiotidis MI. Reactive oxygen species (ROS) – induced genetic and epigenetic alterations in human carcinogenesis. Mutat Res. 2011;711(1–2):167–173. | ||
Smijs TG, Pavel S. Titanium dioxide and zinc oxide nanoparticles in sunscreens: focus on their safety and effectiveness. Nanotechnol Sci Appl. 2011;4:95–112. | ||
Banerjee K, Thiagarajan P. A review of titanium dioxide nanoparticles – synthesis, applications and toxicity concerns. Nanosci Nanotechnol-Asia. 2014;4(2):132–143. | ||
Moezzi A, McDonagh AM, Cortie MB. Zinc oxide particles: synthesis, properties and applications. Chem Eng J. 2012;185:1–22. | ||
Carmichael J, DeGraV WG, Gazdar AF, Minna JD, Mitchell JB. Evaluation of a tetrazolium-based semiautomated colorimetric assay: assessment of chemosensitivity testing. Cancer Res. 1987;47(4):936–942. | ||
Achwal CW, Iyer CS, Chandra HS. Immunochemical evidence for the presence of 5mC, 6mA and 7mG in human, Drosophila and mealy bug DNA. FEBS Lett. 1983;158(2):353–358. | ||
Deobagkar DD, Panikar C, Rajpathak SN, Shaiwale NS, Mukherjee S. An immunochemical method for detection and analysis of changes in methylome. Methods. 2012;56(2):260–267. | ||
Kelkar A, Deobagkar DD. A novel method to assess the full-genome methylation profile using monoclonal antibody combined with the high throughput based microarray approach. Epigenetics. 2009;4(6):415–420. | ||
Sayes CM, Reed KL, Warheit DB. Assessing toxicity of fine and nanoparticles: comparing in vitro measurements to in vivo pulmonary toxicity profiles. J Toxicol Sci. 2007;97:163–180. | ||
Sies H. Glutathione and its role in cellular functions. Free Radic Biol Med. 1999;27(9–10):916–921. | ||
Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–254. | ||
Groote ML, Verschure PJ, Rots MG. Epigenetic editing: targeted rewriting of epigenetic marks to modulate expression of selected target genes. Nucl Acids Res. 2012;40(21):10596–10613. | ||
Robertson KD, Wolffe AP. DNA methylation in health and disease. Nat Rev Genet. 2000;1(1):11–19. | ||
Halušková J. Epigenetic studies in human diseases. Folia Biol (Praha). 2010;56(3):83–96. | ||
Deobagkar D. DNA methylation and toxicogenomics. In: Sahu SC, editor. Toxicology and Epigenetics. Chichester, UK: John Wiley & Sons, Ltd; 2012. | ||
Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8(4):253–262. | ||
Funaki S, Nakamura T, Nakatani T, et al. Global DNA hypomethylation coupled to cellular transformation and metastatic ability. FEBS Lett. 2015;589(24):4053–4060. | ||
Baan RA. Carcinogenic hazards from inhaled carbon black, titanium dioxide, and talc not containing asbestos or asbestiform fibers: recent evaluations by an IARC Monographs Working Group. Inhal Toxicol. 2007;19:213–228. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.