Back to Journals » Infection and Drug Resistance » Volume 11
Epidemiological and molecular analysis of avian influenza A(H7N9) virus in Shanghai, China, 2013–2017
Authors Wang SJ, Liu XW, Shen X, Hua XG, Cui L
Received 8 July 2018
Accepted for publication 16 October 2018
Published 22 November 2018 Volume 2018:11 Pages 2411—2424
DOI https://doi.org/10.2147/IDR.S179517
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Sahil Khanna
Seong Jin Wang,1–3 Xue Wei Liu,1 Xiaojuan Shen,1,2 Xiu Guo Hua,1,2 Li Cui1,2
1Department of Animal Science, Shanghai Key Laboratory of Veterinary Biotechnology, Shanghai Jiao Tong University, Shanghai 200240, China; 2Department of Animal Science, School of Agriculture and Biology, Shanghai Jiao Tong University, Shanghai 200240, China; 3Animal and Plant Quarantine Agency, Gimcheon 39660, Republic of Korea
Background: Human infections with a novel avian influenza A virus (H7N9) were reported in Shanghai municipality, China, at the beginning of 2013. High-pathogenic avian influenza (HPAI) H7N9 virus emerged in late February 2017 along with existing low-pathogenic avian influenza (LPAI) H7N9 virus, and this has the potential to develop into a pandemic that could be harmful to humans.
Methods: To elucidate the epidemiological characteristics of H7N9-infected cases from 2013 to 2017 in Shanghai, data on the 59 laboratory-confirmed human cases and 26 bird and environmental contamination cases were collected from the WHO website and Food and Agriculture Organization Emergency Prevention System for Animal Health (FAO EMPRES-AH). Full-length sequences of H7N9 viruses that emerged in Shanghai were collected from the Global Initiative on Sharing Avian Influenza Data to analyze the evolutionary and genetic features.
Results: We found that genetically different strains emerged in every epidemic in Shanghai, and most of the circulating H7N9 strains had affinity to human-type receptors, with the characteristics of high-virulence and low-pathogenic influenza viruses. Furthermore, our findings suggest that the Shanghai chicken strains are closely related to the HPAI H7N9 virus A/Guangdong/17SF003/2016, indicating that this viral strain is of avian origin and generated from the LPAI H7N9 viruses in Shanghai. The gradual decrease in H7N9 human infection in Shanghai was probably due to the control measures taken by the Shanghai government and the enhanced public awareness leading to a reduced risk of H7N9 virus infection. However, LPAI H7N9 viruses from poultry and environmental samples were continually detected in Shanghai across the epidemics, increasing the risk of new emerging H7N9 outbreaks.
Conclusion: It is important to consistently obtain sufficient surveillance data and implement prevention measures against H7N9 viruses in Shanghai municipality.
Keywords: epidemiology, H7N9 virus, phylogenetic tree, molecular evolutionary study
Introduction
Since the first reports of the outbreak of human infection caused by a novel avian influenza A H7N9 virus [A(H7N9) virus] in Shanghai, China, in March 2013, there have been a total of five epidemics up to 2017. As of 31 December 2017, a total of 1,625 human infections with 623 deaths have been reported, with a mortality rate of approximately 38%.1 Direct or indirect contact with poultry may result in human infection with A(H7N9) virus.2 Before the fifth epidemic, A(H7N9) viruses circulating among poultry in China had been categorized as low-pathogenic avian influenza (LPAI), and the infected poultry showed no symptoms.3 Since poultry carrying the LPAI A(H7N9) virus rarely show signs of sickness, resulting in the virus not being detectable in poultry plants or live poultry markets (LPMs), it is difficult to monitor and prevent the spread of the virus, which increases the risk that the LPAI A(H7N9) viruses in poultry could promote human infections and deaths.2,4 During February 2017, in the fifth epidemic, the appearance of a high-pathogenic avian influenza (HPAI) A(H7N9) virus infection in humans was first reported in Guangdong province.5 Since then, humans and poultry have suffered from infections across China resulting from the outbreak of the HPAI and LPAI A(H7N9) viruses.
A recent study suggests that the HPAI H7N9 virus originated and evolved from the insertion of multiple basic amino acids into the hemagglutinin (HA) protein cleavage site of the LPAI A(H7N9) virus. Then, LPAI A(H7N9) or A(H9N2) viruses reassorted with the HPAI A(H7N9) viruses, resulting in multiple different genotypes.2 The sudden increase in the number of human infections during the fifth epidemic is likely to be associated with the broad spread of the LPAI and HPAI A(H7N9) viruses, which have high levels of genetic variations, host adaptations, and drug-tolerant mutations.6 Owing to the sharp increase in the numbers of human infections with A(H7N9) virus and the outbreaks of HPAI A(H7N9) during the fifth epidemic, there is growing concern regarding the possibility of a pandemic, which has attracted the public’s attention.5
Although some groups of limited human-to-human transmission have been reported,7 there is no evidence of sustained human-to-human transmission of the A(H7N9) virus.8 A(H7N9) and seasonal A(H3N2) virus co-infection, which was first reported at the beginning of 2013, shows that people could be mixing vessels and this could promote human-to-human transmission.9 When human infections with A(H7N9) virus do occur, they are associated with serious illness and high fatality rates. Continuous surveillance is important to identify molecular substitutions in the virus that may have epidemiological characteristics, including increased poultry-to-human transmission and antiviral drug resistance.10
Shanghai is an international city located at latitude 31°14’N, longitude 121°29’E, in front of the Yangtze River delta area. This area is on the East Asian Australian migration flyway, suggesting the probable contribution of wild birds to the emergence of A(H7N9) viruses because migratory shorebirds are a natural reservoir for avian influenza virus.11–13 Several coastal wetland areas, which are significant congregation sites for migratory birds, are also located in Shanghai, increasing the risk of the emergence of novel influenza viruses.14 It should be noted that the first case of human infection with the A(H7N9) virus was reported in Shanghai early in 2013. Although many studies have been performed on A(H7N9) infection, the epidemiological and evolutionary relationships between A(H7N9) viruses circulating in Shanghai remain unclear.
Here, we conducted an epidemiological investigation of A(H7N9) outbreaks in Shanghai and analyzed the A(H7N9) virus strains originating in Shanghai from 2013 to 2017. The phylogenetic and genetic characteristics of A(H7N9) viruses separated from Shanghai were investigated to reveal the evolutionary history of A(H7N9) viruses in Shanghai, along with a detailed discussion on the molecular evolutionary dynamics of the virus. Our findings provide insight into the outbreaks of A(H7N9) virus in Shanghai.
Methods
Epidemiological analysis
The epidemiological study included all laboratory-confirmed cases of human infection with A(H7N9) virus as reported by the WHO website and the Shanghai Municipal Commission of Health and Family Planning (SHMCHFP) from 1 January 2013 to 31 December 2017. Avian influenza reports published by the Department of Health, Government of the Hong Kong Special Administrative Region (Hong Kong SAR) were collected as a supplementary source to identify the cases with human infection. Collected information included the month of onset and the patient’s age and gender. For eight human infection cases with data on the onset of illness missing, the date of the onset was calculated by subtracting 10 days from the WHO reported date. We also collected information on preventive measures implemented by the Shanghai municipal government. We defined the first epidemic as being from January 2013 to September 2013, as defined by the WHO, and subsequent epidemics as being from October to September of the following years.
In total, 59 confirmed A(H7N9) human cases were identified and included in this research. We generated the epidemic curve using the month of illness onset of reported cases and also described the epidemiological characteristics of reported cases to illustrate five epidemics between 2013 and 2017. Birds or environmental cases contaminated with A(H7N9) virus in Shanghai were also downloaded from the Food and Agriculture Organization Emergency Prevention System for Animal Health (FAO EMPRES-AH). The observation dates of the bird and environmental contamination cases were collected to identify the outbreak of A(H7N9) infection in Shanghai.
Sequence alignment and molecular evolutionary analysis
From the Global Initiative on Sharing Avian Influenza Data (GISAID) Database, we downloaded the full-length sequences of the A(H7N9) virus identified during 2013–2017 from Shanghai, as well as vaccine candidates recommended by the WHO [A/Guangdong/17SF003/2016(17SF003), A/HongKong/125/2017(HK125), A/Shanghai/2/2013(SH2), and A/Anhui/1/2013(AN1)]. As a result, HA (n=59), neuraminidase (NA) (n=77), polymerase acidic protein (PA) (n=56), polymerase basic protein 2 (PB2) (n=56), polymerase basic protein 1 (PB1) (n=49), matrix protein (MP) (n=66), nucleoprotein (NP) (n=55), and non-structural protein (NS) (n=66) gene sequences of H7N9 virus were obtained. Using the CLUSTAL W program,15 we performed multiple sequence alignments to find the consensus arrangement and to identify the amino acid residues of interest in the proteins. Genetic distances of the selected strains were calculated by pairwise distance calculations. A phylogenetic tree based on the amino acid sequence of HA or NA in A(H7N9) viruses was constructed for each protein by the neighbor-joining (NJ) method and maximum-likelihood (ML) method using MEGA version 7.016 with 1,000 bootstrap replicates. In the ML method, the Jones–Taylor–Thornton (JTT) model was applied as a substitution model. The A(H7N9) strain A/turkey/Minnesota/1/1988 (GISAID accession number EPI457629) was used as an outgroup to root the tree. The trees were drawn and measured, with branch lengths scaled to the number of mutations per site.
Results
Epidemiological analysis
As shown in Figure 1, five epidemics of A(H7N9) outbreak have swept through the Shanghai municipality. There were seasonal changes in the number of infected cases, and most humans were infected with A(H7N9) virus during November to March. From January 2013 to December 2017, a total of 59 human cases of A(H7N9) infection was reported in Shanghai (Table 1); at least 20 (34%) of these infections resulted in death (Figure 2B). The first epidemic of human infections with the A(H7N9) virus began in early 2013 in mainland China, followed by four consecutive epidemics with 33, eight, eight, four, and six laboratory-confirmed cases. Regarding the occurrence of bird and environmental cases, 20 cases were reported in the first epidemic, one case in the second epidemic, two cases in the third epidemic, one case in the fourth epidemic, and two cases in the fifth epidemic. In both human and bird and environmental cases, more infections were reported during the first epidemic than during the last four epidemics put together (Figure 2C). Given that each epidemic combined into a single period to minimize the risk of the small sample size, 80% of infections occurred in males (Figure 2A), and the median age was 61.5 years (range, 2–89 years). The median age in the first epidemic was 68 years, much older than that observed during the entire period; in the fifth epidemic, the median age was 51.5 years, which was much lower than that observed in other epidemics (Figure 2D). We were unable to analyze correlation among epidemics, and the estimates as presented above should be regarded as indicative because of the small number of cases of human infection in the second to fifth epidemics.
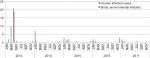  | Figure 1 Confirmed cases of avian influenza A(H7N9) virus infections in humans, birds, and the environment in Shanghai reported to the WHO and Food and Agriculture Organization (n=85), by month of illness onset, 2013–2017. Notes: Origin: Publicly released infections in the Disease Outbreak News (http://www.who.int/csr/don/en/) or avian influenza report issued by the Department of Health, Government of the Hong Kong Special Administrative Region (http://www.chp.gov.hk/en/index.html). |
Molecular homology analysis of amino acid sequences
Homology analyses of amino acid sequences of selected strains indicated that the variation between the strains showed diversity at the amino acid level up to 4.6%, and 5.5% was based on the surface HA and NA genes, with variable differences being observed based on internal gene sequences. Comparison of the internal gene sequences of the selected strains revealed amino acid divergences up to 2.5% (PB2), 2.4% (PB1), 3.6% (PA), 4.9% (NP), 7.9% (MP), and 13.3% (NS) (Table 2). The two recent A(H7N9) vaccine candidates had higher amino acid divergences of HA and NA proteins from the Shanghai strains than the vaccine candidates of 2013. The HA and NA surface proteins of Shanghai strains, with four different vaccine candidates, shared 95.1–98.9% and 96.1–98.9% (HK125), 94.9–97.8% and 94.7–97.6% (17SF003), 96.7–100% and 96.3–100% (SH2), and 96.7–100% and 96.5–100% (AN1), respectively, at the amino acid level (Table 3).
Phylogenetic study of the HA and NA proteins in Shanghai strains
A total of 59 HA and 77 NA full-length sequences of A(H7N9) strains were obtained from the GISAID database (http://www.gisaid.org) to generate the phylogenetic tree. Both the NJ trees and ML trees, based on HA segments, showed that they shared similar topologies, and all of the H7N9 strains formed a closely related lineage except for four strains, namely A/duck/Shanghai/SD015(SD015), A/duck/Shanghai/SD016(SD016), A/Shanghai/1/2013(SH1), and A/Shanghai/05/2013(SH05), when the tree was rooted to strain A/turkey/Minnesota/1988. These four strains had nodes and branches distinct from other strains in both the ML and NJ methods. It was also observed that the A(H7N9) WHO vaccine candidates were closely related to A/chicken/Shanghai/S4100/2015 (Figure 3A and B). The phylogenetic trees of NA segments of Shanghai strains, as shown in Figure 4A and B, revealed that the ML and NJ trees were broadly similar in topology and the strains formed several clades. Two vaccine candidates recommended by the WHO, 17SF003 and HK125, were clustered with the Shanghai chicken strains of 2014 and 2015 to form a clade. As a result, we inferred that the source of HPAI A(H7N9) viruses came from poultry. Based on the two methods of phylogenetic analysis of HA and NA protein, SH05, SD015, and SD016 produced a branch distinct from the other Shanghai strains and were located closest to the common ancestor.
Key amino acid substitutions of A (H7N9) viruses in Shanghai municipality
The genetic characteristics of Shanghai strains that were associated with host range, receptor binding specificity, and antiviral tolerance were analyzed. We found that 91% (50/55) and 92.7% (51/55) of strains contained G186V (H3 numbering) and Q226L/I mutations, respectively, in the HA protein, which may enhance the human receptor binding affinity of the virus.17 SH05, SD015, and SD016 had no mutations with 186G and 226Q, while A/chicken/Shanghai/S1410/2013 had G186V mutations with 226Q, and SH1 and A/Minhang/S01/2013 had Q226L/I mutations with 186G (Tables 4 and 5).
The HA of all strains from Shanghai included a single arginine at the HA cleavage site, as shown in Table 4, suggesting that Shanghai strains belong to the LPAI viruses. Only the vaccine candidate strain 17SF003 had an insert of multiple basic amino acid residues at the HA cleavage site (PKRKRTAR/G) in this survey (Table 5). R292K mutations of NA protein related to antiviral tolerance occurred in only three strains, including SH1. A key five-amino-acid deletion in the NA protein at positions 69–73 has been associated with its increased virulence.18 The Shanghai strains analyzed in this study had a five-amino-acid (positions 69–73) deletion in the NA stalk region, except for SH05, SD015, and SD016.
Several mutations associated with mammalian adaptation were generated in the internal protein of Shanghai strains. The E627K and D701N mutations in the PB2 protein, and V100A and K356R mutations in PA, are regarded as essential for mammalian adaptation of the avian influenza virus.19–21 E627K mutations in the PB2 protein occurred in a total of 20 viruses, including A/tree sparrow/Shanghai/01/2013. The D701N mutation was only observed in A/Shanghai/PD-02/2014 (Table 5). The PA genes from 52 viruses were also analyzed. While V100A mutations were presented in 42 strains, the K356R substitution was observed in the majority of the strains except for in SD015 and SD016 (Table 5). I368V mutations in PB1, which were related to increased transmission in ferrets, were observed, except for in SH1 and A/Shanghai/11/2013 (Table 5).
All Shanghai viruses as well as WHO-recommended vaccine candidates had P42S and N205S substitutions, indicating their enhanced virulence in mice and altered antiviral response (Table 5). However, the D92E mutation in the NS protein related to increased virulence and/or cytokine resistance was found only in SH05. In the genes of the SH05 strain, we observed some mammalian adaptation signatures such as the I368V mutation in PB1, K356R in PA, and avian-like signatures including G186 and Q226, which retained the 69–73 amino acids in the stalk region in HA and V100 in PA. Ren et al21 conjectured that SH05 had more avian influenza-like characteristics and was genetically distinct from other human influenza-like strains, representing a precursor of A(H7N9) subtypes co-circulating among humans. It was observed that SD015 and SD016 had more avian influenza-like characteristics than SH05.
Discussion
The A(H7N9) virus that emerged in 2013 originated from the reassortment between the A(H7N3) virus HA gene in Zhejiang ducks and A(H7N9) virus NA gene in Korean wild birds, adopting six internal protein genes of the A(H9N2) virus in Beijing bramblings.11 HA assists the virus in attaching to and penetrating the host cell via surface receptors, while NA catalyzes the cleavage of sialic acid residues on the surface receptor, which is believed to play important roles in influenza infection. It is noteworthy that a single amino acid mutation of the receptor binding site in HA may contribute to the change in receptor binding specificity.22 The HA protein harbored G186V and Q226L (H3 numbering) mutations and has previously been identified as being related to a switch in receptor specificity from poultry to human types.23 Through the tuning procedure of enhancing the affinity to human-type receptors, the virus acquired dual receptor binding capacity.24 A total of 50 and 51 viruses, including most of the poultry-derived strains, in this study had the G186V and Q226L mutations, respectively, in their HA segments, indicating that most of the Shanghai strain possessed human virus-like receptor preference.
The NJ method is a well-known distance method, and the ML method is the most commonly used analysis in phylogenetic tree construction.25 We applied the two methods and compared the results of the constructed trees in this study. We observed that these two trees were broadly congruent with the topology and shared some internal nodes and branches near the ancestral node in the analysis of HA and NA proteins. The ancestral HA and NA genes of the A(H7N9) virus were observed in SH05, SD015, and SD016, as these viruses were phylogenetically close to the common ancestor. While a five-amino-acid deletion (positions 69–73) in the HA stalk region is considered a characteristic feature of wild aquatic bird viruses adapted to terrestrial birds, these three viruses contain a long stalk in NA, 186G, and 226Q in HA, indicating that these viruses are associated with aquatic bird viruses. Wang et al24 concluded that early viruses such as these strains could spread to terrestrial birds and acquire a five-amino-acid deletion (QISNT) in the NA stalk region, producing terrestrial poultry-adapted SH1-like viruses. However, how the SH05 without both G186V and Q226L and the strains with either G186V or Q226L could still infect humans requires further investigation.
It is important to note the phylogenetic subgroup consisting of SD015 and SD016, because the domestic duck could be a key intermediate host for viral reassortment and ultimately produce A(H7N9) virus. It was observed that SD015 and SD016 have different genetic characteristics that appeared in the third epidemic. A duck transferring H7N9 into its environment could potentially lead to the infection of domestic poultry.26 Therefore, it is necessary to strengthen the continuous surveillance of birds and environmental samples.
As shown in Figure 3, the WHO-recommended HPAI A(H7N9) vaccine candidate 17SF003 was rooted from the Shanghai strains. This finding shows that the HPAI H7N9 virus is of avian origin, stemming from the LPAI A(H7N9) viruses in Shanghai. Systemic virus replication in poultry and mammals could be facilitated by the insertion of multiple basic amino acids in the HA cleavage site of the vaccine candidates.27–29 This insertion is considered as a primary virulence factor of A HPAI viruses.30 While no Shanghai strains contain these mutations on HA segments, the insertion of multiple basic amino acids in the cleavage site was observed in the vaccine candidate 17SF003. As stated above, most Shanghai strains possessed the QISNT deletion in the NA protein, which may be associated with its increased virulence. These results suggest that most of the Shanghai strains are low-pathogenic and high-virulence strains.
NA offers an excellent target for antiviral drug development. R292K substitution in NA has been reported to be resistant to NA inhibitor antiviral drugs.31 In the current study, only 4% of the Shanghai strains (3 of 73 isolates, only including the isolates in 2013) contained the antiviral resistance R292K mutations, and no substitutions were observed in the environmental strains. These findings show that NA inhibitor antiviral drugs may provide a valid remedy for A(H7N9)-infected patients in Shanghai.
The polymerase enzyme complex essential for viral replication is composed of PB2, PB1, and PA genes. Mutations of these genes are linked with increased virulence and animal-to-human transmissibility.20,32 Based on the different genetic characteristics of the surface proteins and internal proteins in H7N9 viruses, it appears that the genetically new H7N9 strains have emerged in every epidemic in Shanghai, posing a risk to public health. Although only non-sustained and limited transmission was detected, a sharp increase in human infections and a higher pathogenicity in chickens were reported in the recent epidemics.33,34 The A(H7N9) viruses from 32 human cases and 57 poultry or environmental samples were found to be HPAI viruses, but there have been no reports of an HPAI A(H7N9) outbreak in Shanghai.1
Geographically, A(H7N9) virus infection extended from eastern to western China across the epidemics. In addition, an increasing number of cases of human infections was observed in the fifth epidemic compared to any of the previous epidemics in mainland China.35 In contrast, 56% of the human cases, with a 48% fatality rate, were reported in the first epidemic in Shanghai. A relatively small number of cases of human infection were observed from the second to fifth epidemics in Shanghai. Shen et al36 discovered that during the first epidemic in Shanghai, late antiviral treatment and a long duration of viral shedding resulting from deficient knowledge among the general public of the A(H7N9) virus were more likely to be associated with a fatal outcome. Cases of A(H7N9) infection gradually decreased from the first epidemic to the fourth epidemic, and only four fatal cases and 28 total cases were observed in Shanghai in the last four epidemics. This decrease in the number of A(H7N9) infection cases and the low fatality rate may be associated with enhanced public awareness, which resulted in a reduced risk of A(H7N9) virus infection,3 along with continuous surveillance and prevention measures.
After the first human infection with the A(H7N9) virus, the Shanghai government implemented enhanced prevention and control programs, including the closure of the LPMs, slaughtering poultry, and investigation of environmental samples, as well as implementing an emergency monitoring plan.37 Although this promoted awareness of risk reduction to decrease A(H7N9) viral infections in the people of Shanghai, the LPAI A(H7N9) viruses in poultry and environmental samples have been continually detected in Shanghai across the epidemics, enhancing the risk of silent epidemics and occurrences leading to sporadic human infections caused by contact with infected poultry.38
After the first human infection with the A(H7N9) virus in Shanghai, all 464 LPMs were temporarily closed on 6 April 2013. The LPMs were cleaned and disinfected, and relevant authorities in the neighboring provinces were informed that vehicles should not transport live poultry to Shanghai municipality. He et al suggested that the poultry in LPMs may have infected humans, with or without direct contact.39 In January 2014, the Shanghai municipal government issued a ban stating that the LPMs would be regularly closed from every Chinese New Year to April. After the LPMs were closed on the 2014 Chinese New Year, no cases were reported in the second epidemic. However, the closure of LPMs may not be the deciding factor in preventing the occurrence of A(H7N9) in Shanghai because a few of the remaining A(H7N9) cases occurred during the ban period from 2015 to 2017. The start dates of the LPM closures in Shanghai were 29 February 2015, 8 February 2016, and 28 January 2017.
Shanghai is one of the least productive municipalities for poultry in China. Live poultry production in Shanghai gradually decreased from 2013 (26.41 million) to 2015 (19.43 million). Also, poultry consumption in Shanghai is approximately 130 million annually, of which 20% is from Shanghai municipality and 80% is mainly from Jiangsu, Anhui, and Zhejiang, indicating that most of the poultry consumed in Shanghai is imported from other provinces.40 Therefore, when investigating the outbreaks of poultry-related diseases in Shanghai, it is necessary to be aware of the poultry imported from other provinces. The closure of LPMs may have facilitated the movement of LPAI A(H7N9) virus-infected poultry to the Shanghai district during the ban period. The poultry trade and transportation routes could facilitate the silent spread of LPAI A(H7N9) virus.3 HPAI A(H7N9) viruses may also spread to other provinces via bird migratory flyways.2
According to the routine environmental surveillance in Shanghai, H7N9-positive samples dramatically decreased in the second to fifth epidemics compared to the first epidemic but were continuously detected across the epidemics, increasing the risk of new emerging H7N9 outbreaks. Also, migratory birds and poultry transportation could facilitate LPAI and HPAI A(H7N9) outbreaks. Therefore, it is necessary to focus on continuous prevention and surveillance for the A(H7N9) virus in Shanghai municipality. Because China is composed of large provinces, each of which has a wide range of population, climate, and environment, province-based quarantine and prevention could be more effective than nationwide policies in controlling infectious disease. WHO and Chinese experts assessed that quarantine measures conducted by the Shanghai government against A(H7N9) infection were considered to be appropriate and effective41 and also could provide a good example for A(H7N9) control and prevention in other regions.
Conclusion
In summary, we found that genetically new A(H7N9) strains emerged in every epidemic in Shanghai, and most of the emergent strains had affinity for human-type receptors. The A(H7N9) viruses circulating within Shanghai belong to high-virulence and low-pathogenic influenza viruses, considering their NA deletion (positions 69–73) and the insertion of single basic amino acids within the HA cleavage site. In addition, it is likely that NA inhibitor antiviral drugs may be a valid remedy for A(H7N9)-infected patients in Shanghai, based on the fact that only three Shanghai viruses during the first epidemic contained the R292K mutations and no environmental or bird samples including the mutations had yet been reported. This study also indicates that the HPAI A(H7N9) virus that emerged in the fifth epidemic is of avian origin because it is phylogenetically closely related to a Shanghai chicken strain. The improved public awareness of A(H7N9) pandemic risk,3 as well as several surveillance and control measures, may have contributed to the gradual decrease in A(H7N9) outbreaks in Shanghai. However, LPAI A(H7N9) viruses found in poultry and environmental samples were continually detected in Shanghai across the epidemics, increasing the risk of new emerging A(H7N9) outbreaks. Also, LPAI or HPAI A(H7N9) viruses could spread through poultry transportation and bird migration pathways.2,3 It is important to take consistent and sufficient surveillance and prevention measures against H7N9 viruses in Shanghai municipality.
There are some limitations to this study. First, the human cases of A(H7N9) infection during the second to fifth epidemics in Shanghai numbered only eight, eight, four, and six, respectively. Because the relatively small number of confirmed cases during the second to fifth epidemics makes it difficult to have a large enough sample size to generate meaningful statistics and find statistically significant differences in the distributions of A(H7N9) outbreaks, our estimates for fatality, median age, and gender distribution of the study could be biased. Therefore, it is recommended that well-designed pilot studies, with small sample sizes, or pooled analyses with confirmed cases from other regions, are undertaken to find significant differences in future research. In addition, some sequences or cases have been missed because they were not available from the database used in this study. As the study cases are not representative of the entire H7N9 group, the results could be skewed. Second, for the cases infected with H7N9, access to updated information, including mortality cases, was limited. Further studies including additional epidemiological and clinical information are required for a better understanding of H7N9 human infection in Shanghai.
Acknowlegdment
This work was financially supported by the National Key Research and Development Program of China (2017YFC 1200202) and the Shanghai Agriculture Applied Technology Development Program, China (Grant No. HuNongKe [2017] 1-11).
Disclosure
The authors report no conflicts of interest in this work.
References
Food and Agricultural Organization of the United Nations [webpage on the Internet]. H7N9 situation update; 2018. Available from: http://www.fao.org/ag/againfo/programmes/en/empres/H7N9/situation_update.html. Accessed June 4, 2018. | ||
Yang L, Zhu W, Li X, et al. Genesis and Spread of Newly Emerged Highly Pathogenic H7N9 Avian Viruses in Mainland China. J Virol. 2017;91(23). | ||
Wang X, Jiang H, Wu P, et al. Epidemiology of avian influenza A H7N9 virus in human beings across five epidemics in mainland China, 2013-17: an epidemiological study of laboratory-confirmed case series. Lancet Infect Dis. 2017;17(8):822–832. | ||
Peiris JS, Cowling BJ, Wu JT, et al. Interventions to reduce zoonotic and pandemic risks from avian influenza in Asia. Lancet Infect Dis. 2016;16(2):252–258. | ||
Zhang F, Bi Y, Wang J, et al. Human infections with recently-emerging highly pathogenic H7N9 avian influenza virus in China. J Infect. 2017;75(1):71–75. | ||
Quan C, Shi W, Yang Y, et al. New Threats from H7N9 Influenza Virus: Spread and Evolution of High- and Low-Pathogenicity Variants with High Genomic Diversity in Wave Five. J Virol. 2018;92(11). | ||
Qin Y, Horby PW, Tsang TK, et al. Differences in the Epidemiology of Human Cases of Avian Influenza A(H7N9) and A(H5N1) Viruses Infection. Clin Infect Dis. 2015;61(4):563–571. | ||
Iuliano AD, Jang Y, Jones J, et al. Increase in Human Infections with Avian Influenza A(H7N9) Virus During the Fifth Epidemic - China, October 2016-February 2017. MMWR. 2017;66(9):254–255. | ||
Zhu Y, Qi X, Cui L, Zhou M, Wang H. Human co-infection with novel avian influenza A H7N9 and influenza A H3N2 viruses in Jiangsu province, China. Lancet. 2013;381(9883):2134. | ||
Kile JC, Ren R, Liu L, et al. Update: Increase in Human Infections with Novel Asian Lineage Avian Influenza A (H7N9) Viruses During the Fifth Epidemic – China, October 1, 2016–August 7, 2017. MMWR. 2017;66(35):928–932. | ||
Gao R, Cao B, Hu Y, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368(20):1888–1897. | ||
Capua I, Alexander DJ. Avian influenza: recent developments. Avian Pathol. 2004;33(4):393–404. | ||
Alvarez P, Mattiello R, Rivailler P, et al. First isolation of an H1N1 avian influenza virus from wild terrestrial non-migratory birds in Argentina. Virology. 2010;396(1):76–84. | ||
Bai Q, Chen J, Chen Z, Zeng Q. Identification of coastal wetlands of international importance for waterbirds: a review of China Coastal Waterbird Surveys 2005–2013. Avian Res. 2015;6(1):12. | ||
Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. | ||
Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33(7):1870–1874. | ||
Xiong X, Martin SR, Haire LF, et al. Receptor binding by an H7N9 influenza virus from humans. Nature. 2013;499(7459):496–499. | ||
Munier S, Larcher T, Cormier-Aline F, et al. A genetically engineered waterfowl influenza virus with a deletion in the stalk of the neuraminidase has increased virulence for chickens. J Virol. 2010;84(2):940–952. | ||
van Riel D, Munster VJ, de Wit E, et al. H5N1 Virus Attachment to Lower Respiratory Tract. Science. 2006;312(5772):399. | ||
Li Z, Chen H, Jiao P, et al. Molecular basis of replication of duck H5N1 influenza viruses in a mammalian mouse model. J Virol. 2005;79(18):12058–12064. | ||
Ren L, Yu X, Zhao B, et al. Infection with possible precursor of avian influenza A(H7N9) virus in a child, China, 2013. Emerg Infect Dis. 2014;20(8):1362–1365. | ||
Rogers GN, Paulson JC, Daniels RS, Skehel JJ, Wilson IA, Wiley DC. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature. 1983;304(5921):76–78. | ||
Imai M, Watanabe T, Hatta M, et al. Experimental adaptation of an influenza H5 haemagglutinin (HA) confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature. 2012;486(7403):420–428. | ||
Wang D, Yang L, Gao R, et al. Genetic tuning of the novel avian influenza A(H7N9) virus during interspecies transmission, China, 2013. Euro Surveill. 2014;19(25). | ||
Lam TT, Hon CC, Tang JW. Use of phylogenetics in the molecular epidemiology and evolutionary studies of viral infections. Crit Rev Clin Lab Sci. 2010;47(1):5–49. | ||
Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56(1):152–179. | ||
Schrauwen EJ, Herfst S, Leijten LM, et al. The multibasic cleavage site in H5N1 virus is critical for systemic spread along the olfactory and hematogenous routes in ferrets. J Virol. 2012;86(7):3975–3984. | ||
Shinya K, Makino A, Hatta M, et al. Subclinical brain injury caused by H5N1 influenza virus infection. J Virol. 2011;85(10):5202–5207. | ||
Shinya K, Makino A, Tanaka H, et al. Systemic dissemination of H5N1 influenza A viruses in ferrets and hamsters after direct intragastric inoculation. J Virol. 2011;85(10):4673–4678. | ||
Horimoto T, Kawaoka Y. Reverse genetics provides direct evidence for a correlation of hemagglutinin cleavability and virulence of an avian influenza A virus. J Virol. 1994;68(5):3120–3128. | ||
McKimm-Breschkin J, Trivedi T, Hampson A, et al. Neuraminidase sequence analysis and susceptibilities of influenza virus clinical isolates to zanamivir and oseltamivir. Antimicrob Agents Chemother. 2003;47(7):2264–2272. | ||
Sha J, Chen X, Ren Y, et al. Differences in the epidemiology and virology of mild, severe and fatal human infections with avian influenza A (H7N9) virus. Arch Virol. 2016;161(5):1239–1259. | ||
Zhou L, Ren R, Yang L, et al. Sudden increase in human infection with avian influenza A(H7N9) virus in China, September-December 2016. Western Pac Surveill Response J. 2017;8(1):6–14. | ||
Li Q, Zhou L, Zhou M, et al. Epidemiology of human infections with avian influenza A(H7N9) virus in China. N Engl J Med. 2014;370(6):520–532. | ||
Su S, Gu M, Liu D, et al. Epidemiology, Evolution, and Pathogenesis of H7N9 Influenza Viruses in Five Epidemic Waves since 2013 in China. Trends Microbiol. 2017;25(9):713–728. | ||
Shen Y, Lu H, Qi T, et al. Fatal cases of human infection with avian influenza A (H7N9) virus in Shanghai, China in 2013. Biosci Trends. 2015;9(1):73–78. | ||
Shanghai Municipal Commission of Health and Family Planning. [Prevention and control measures of Human infection with H7N9 influenza virus in Shanghai municipality]. Available from: http://www.wsjsw.gov.cn/xwfb/20180525/35470.html. Accessed November 13, 2018. Chinese. | ||
Kim SM, Kim YI, Pascua PN, Choi YK. Avian Influenza A Viruses: Evolution and Zoonotic Infection. Semin Respir Crit Care Med. 2016; 37(4):501–511. | ||
He Y, Liu P, Tang S, et al. Live poultry market closure and control of avian influenza A(H7N9), Shanghai, China. Emerg Infect Dis. 2014;20(9):1565–1566. | ||
Shanghai Municipal Agricultural Commission. Information abstract (internal data). Shanghai Agricul Netw. 2013;303(7):1–8. | ||
The Central People’s Government of the People’s Republic of China. [Shanghai’s response to the H7N9 epidemic is worthy of recognition]; 2013. Available from: http://www.gov.cn/jrzg/2013-04/23/content_2386475.htm. Accessed November 13, 2018. Chinese. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.








