Back to Journals » Journal of Inflammation Research » Volume 15
Emerging Roles of Post-Translational Modifications in Skin Diseases: Current Knowledge, Challenges and Future Perspectives
Received 14 September 2021
Accepted for publication 12 November 2021
Published 11 February 2022 Volume 2022:15 Pages 965—975
DOI https://doi.org/10.2147/JIR.S339296
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Luting Yang, Yaping Yan
Key Laboratory of the Ministry of Education for Medicinal Resources and Natural Pharmaceutical Chemistry, National Engineering Laboratory for Resource Development of Endangered Crude Drugs in Northwest of China, College of Life Sciences, Shaanxi Normal University, Xi’an, People’s Republic of China
Correspondence: Luting Yang; Yaping Yan, Email [email protected]; [email protected]
Abstract: Post-translational modifications (PTMs) of proteins represent as a key step in regulating their biological functions and dynamic interaction with other players. This process is fine-tuned by a myriad of enzymes named “writers, readers and erasers” whose actions are precisely controlled. Either the mutation, aberration in the expression of the aforementioned enzymes or their substrates have shown to participate in the pathogenesis of various skin diseases such as melanoma, vitiligo, psoriasis, eczema, atopic dermatitis and inherited dermatological diseases. It is becoming increasingly clear that key transcriptional factors, inflammation-related molecules are prone to PTMs. Despite their importance in regulating key processes including inflammation, keratinocyte apoptosis, proliferation and differentiation, PTMs have received less attention due to the challenges involved. Here in this review we summarize the role of the most common types and the newly discovered PTMs, including acetylation, glycosylation, citrullination, PARylation and sumoylation in dermatoses and surveys the recent progress in PTM-based therapeutic approaches in skin diseases.
Keywords: post-translational modification, skin diseases, molecular mechanism, therapeutics
Introduction
Post-translational modification (PTM) is the covalent adding of a functional group to the protein substrates. After translation, the majority of proteins undergo diverse PTMs to maintain their structure, stability and interactions with other players in the complex biological systems. Among the 300 identified forms of PTMs, the most common types of PTMs include ubiquitination, phosphorylation, glycosylation, acetylation and PARylation. These processes are fine-tuned by a myriad of enzymes named “writers, readers and erasers” whose actions are precisely controlled. The interplay and the coordinated network between these PTMs play key roles in regulating protein stability, structure, protein–protein interaction, cell signaling, cell growth, and various biological processes.
The involvement of PTMs in the regulation of skin functions is well documented. Transcriptional factors, signaling molecules and proteins involved in keratinocyte differentiation, proliferation and inflammation are subjected to various PTMs. Consequently, dysregulation in the steps of the aforementioned PTMs and aberrant expression of enzymes involved are related to the pathogenesis of skin cancers, inflammatory skin diseases such as psoriasis, eczema, atopic dermatitis and inherited dermatological diseases. The proteome-wide PTM analysis in these skin diseases helps to clarify the underlying molecular mechanisms and provide new therapeutic targets for the treatment of skin diseases. Herein, we summarized the recent findings concerning the role of the most common types of PTMs in skin diseases, with a focus on acetylation, glycosylation, citrullination, and PARylation. Moreover, we discuss the challenges and prospects for targeting PTMs in the treatment of skin diseases.
Role of Different PTMs in Skin Diseases
Acetylation/Deacetylation
Protein acetylation/deacetylation is a reversible PTM that adds or removes an acetyl group to the amino acid of a protein. This process is catalyzed by two groups of enzymes, lysine acetyltransferases (KAT) and deacetylases (KADC).1 Acetylation or deacetylation of non-histone proteins modifies a series of biological processes including enzymatic activity, inflammation, autophagy, protein–protein interaction and protein localization. Either aberrant expression of acetyltransferases/deacetylases or alteration in the status of acetylation of protein targets are found in various skin diseases such as psoriasis and melanoma.2,3
Psoriasis is an inflammatory skin disease characterized by keratinocyte hyperproliferation and infiltration of immune cells. Upregulation of proliferation-related proteins and inflammatory signaling pathways are key mechanisms accounting for the psoriasis pathogenesis. Predication by PhosphoSitePlus (https://www.phosphosite.org/) suggests these proteins such as Keratin 16, 17, STAT1 and STAT3 could be potentially acetylated on various amino acids, implicating the involvement of acetylation process in the pathogenesis of psoriasis. Sirtuin (SIRT) are a family of NAD+-dependent deacetylases that are composed of seven members in mammals. The abnormal expression of all seven members have been documented by different groups, suggesting a decreased expression of SIRT1-5 and an upregulation of SIRT6 and 7.4 Among these members, SIRT1 is the star molecule that attracts the attention of researchers. The precise mechanism underlying the involvement of SIRTs in psoriasis had been investigated in human cell line and in vivo. There is compelling evidence showing the interaction between SIRT1 and STAT3, a key transcription factor that regulates cell proliferation in psoriasis. STAT3 deacetylation by SIRT1 inhibits its phosphorylation and activation.5 Additionally, another study found that proinflammatory cytokine IFN-γ was responsible for inducing the reduction of SIRT1 and acetylation of STAT3 in psoriatic keratinocytes, which provides an “IFN-γ-SIRT1-STAT3” positive feedback loop in the pathogenesis of psoriasis.6 Later on, application of a SIRT1 activator resveratrol was proven to be effective in the amelioration of Aldara-induced psoriasiform phenotype.7 Collectively, the above findings underlie the crucial role of SIRT1 in the development of psoriasis.
Apart from psoriasis, acetylation is also involved in other skin diseases such as Cutaneous T-cell Lymphomas (CTCLs). TP53, a tumor suppressor gene, represents as a wide-spectrum therapeutic target in cancers. TP53 mutation is found in CTCLs, especially advanced stage of CTCLs patients. P53 is prone to various PTMs such as ubiquitination, acetylation and phosphorylation. Recently, deacetylase HDAC1/8 is reported to interact with p53 and induced its deacetylation and deactivation.8 Herein, the aforementioned deacetylation of p53 represents a novel mechanism accounting for the tumor progression in CTCLs.
Despite the current findings regarding the role of deacetylase SIRTs and HDAC family members, the role of other members in KAT and KDAC has not been elucidated. Further investigations are warranted for deciphering the role of KAT, KDAC in the pathogenesis of skin diseases.
PARylation
ADP-ribosylation, also known as PARylation, is a reversible PTM that adds ADP-ribose moiety to the amino acid residues of the target proteins. This process is catalyzed by PARP members, which function as “writers” of ADP-ribose. PARP contain at least 17 members in humans and are divided into 4 sub-families depending on their functional domains.9 Among the PARP family, PARP1 is the prototypical and founding member, which is the most intensively and extensively studied member. Apart from the well-known function in DNA damage, PARylation is also implicated in inflammation, wound healing, protein stability and cell death.10
Inflammation is a pathological process that occurs in various skin diseases such as psoriasis, eczema and vitiligo. Overwhelming evidence has substantiated the regulatory role of PARPs in inflammation. PARP1 was shown to induce the PARylation of NF-κB, acting as a transcriptional coactivator of NF-κB and promoting the expression of NF-κB target genes.11 Moreover, other substrates such as transcription factor HMGB1 are found to interact with PARP1. PARP1 induced the PARylation and acetylation of HMGB1, promoting the release of this proinflammatory cytokine to the cytoplasm.12 In skin wound healing, which is accompanied with inflammation, it is unraveled that inhibition of PARP1 promotes keratinocyte migration by decreasing the production of pro-inflammatory cytokines.13 Apart from the aforementioned proinflammatory function, PARP could also exert anti-inflammatory functions.14 In psoriasis, PARP1 expression is decreased in the epidermis of psoriatic patients. The PARP1−/− mice exacerbated the IMQ-induced psoriasis-like lesions by increasing the secretion of psoriasis-related proinflammatory cytokines IL-6, IL-17 and IL-23.14
In systemic sclerosis (SSc), which is characterized by immunological alterations in skin and various internal organs,15 a negative role of PARP1 in the regulation of TGF-β/Smad signaling was reported.16 PARP1 expression is decreased in skin of SSc patients due to TGF-β-induced hypermethylation in the promoter of PARP1. Interestingly, Smad3 is found to be a novel target of PARP1, inhibition of PARP1 decreased the PARylation of Smad3, which promotes its phosphorylation and binding to target genes. Herein, the downregulated PARP1 leads to the activation of TGF-β/Smad signaling and contributes to the persistent fibrosis activation in SSc.16
In melanoma, elevated expression of PARP1 is documented by various studies so far. Recently, Kupczyk et al17 found that melanoma patients with high PARP1 expression was frequently located in the neck and head region. Consistently, high PARP1 expression showed correlation with unfavorable clinical outcome such as overall survival and presence of ulceration.18 Experimental studies further highlighted that inhibition of PARP1 expression decreased the migration and invasiveness in melanoma cell lines, supporting the involvement of PARP1 in the development of melanoma.19 PARP1 could potentially regulate the expression of CXCL1 and MITF to promote the growth of melanoma cells.20 Similarly, the upregulation of CXCL1 and MITF in melanoma cell do not appear to depend on the enzymatic activity of PARP1, but seems to rely on the transcriptional co-regulator function of PARP1.20,21 Rodríguez et al22 reported that PARP1 modulates the metastasis and migration of melanomas via regulating the expression of EMT marker vimentin. Table 1 summarizes the role of PARP1 in different skin diseases.
 |
Table 1 The Role of PARP1 in Skin Diseases |
With the development of proteomic technologies, an explosion of understanding on how PARylation impacts not just the process of DNA damage but also various biological functions has been seen. Apart from PARP1, the functions of other members of PARP family remain unknown. What are their functions in regulating the pathogenesis of skin diseases? Is there crosstalk between PARylation and other PTMs, such as acetylation, and how this complex crosstalk mediates skin pathology? The answers of these questions warrant further experimental and clinical investigations which are of importance for developing therapeutic targets for the treatment of skin diseases.
Citrullination
Protein citrullination, also known as deimination, is also a PTM that converts a positively charged arginyl residue into a neutral citrulline residue.23 This process results in a reduction in isoelectric point and 1 Da decrease in mass. As such, the loss of protein charge can affect protein structure, protein–protein interaction, therefore inducing protein conformational change and susceptibility to proteolysis. Although firstly described in 1958, there remains much challenge in identifying the site of citrullination on substrates due to the minor 1 Da change in protein mass. Protein Arginine Deiminases (PAD), which catalyzed this process, are a group of calcium-dependent enzymes.24 There are 5 members of PADs identified in humans, including PAD1, PAD2, PAD3, PAD4 and PAD6. PADs display unique tissue localization and substrate specificity. PAD1 and PAD3 are supposed to be critical for skin physiology and pathology due to their specific expression in the epidermis and hair follicles. Whereas, PAD2 and PAD4 have widespread tissue localization such as tumors, central nervous systems, lungs and inflammatory cells. Due to their widespread expression and the determined three-dimensional structure, PAD2 and PAD4 are the two members that are more extensively studied.25 In physiological conditions, PADs remain inactive due to the very low levels of calcium concentration. In the process of autophagy and keratinocyte differentiation whereby calcium concentration is above the physiological level, PADs are activated. So far, citrullination is reported to be involved in regulating the key cellular processes including apoptosis, inflammation, epidermal differentiation, epidermal barrier function and immune response.23
For deciphering the role of citrullination in different diseases, activators that trigger the activity of PADs except for calcium should be identified. Among the few activators identified, zeta isoform of PKC, PKCζ was previously shown to be an activator of PAD4 which is involved in the release of neutrophil extracellular traps (NET). The possible explanation is that PKCζ could either induce the nuclear translocation or alter the chromatin structure of PAD4 through epigenetic medication.26 Another identified activator is TNF-α. In inflammatory skin diseases such as psoriasis, TNF-α is expressed at high levels both in the serum and skin tissues.27 In multiple sclerosis and rheumatoid arthritis, the elevated level of TNF-α could induce PAD4 nuclear translocation and histone citrullination.28,29 Besides, by library screening, xanthine derivative acefylline was proven to be the potent activator of PAD1 and PAD3. Application of acefylline on skin surface led to an increase in the citrullinated proteins in the stratum corneum.30 Altogether, the aforementioned PKCζ, TNF-α and acefylline are potent activators of PADs which are useful in the investigation of the role of citrullination in different diseases.
The aberration in PAD activity and the subtle change in the citrullination level of substrates are found in several diseases such as psoriasis,31 rheumatoid arthritis,32 cancers33 and neurodegenerative diseases.34 By immunohistochemical staining, PAD4 expression is shown to be elevated in skin carcinomas.33 Also, the mutation of the gene encoding PAD3, PADI3, has been reported in hair diseases such as UHS (Uncombable hair syndrome).35 Psoriasis is the only skin diseases found to be related to PAD1 dysregulation. The absence of deiminated K1 may be one possible explanation for the deranged keratinization process in psoriasis patients.31 Systemic lupus erythematosus is an autoimmune disorder that affect multi-organs, with skin manifestations found in over 85% of patients. Recently, a novel functional role of PAD4 in the pathogenesis of SLE has been characterized.36 A20, a protein disposing a dual role of ubiquitin ligase and deubiquitinase, is known for its function in the negative regulation of NF-κB signaling. Herein, unexpectedly, instead of activating NF-κB activity, genetic alteration in A20 DUB lead to upregulated expression of PAD4. The upregulation of PAD4 contributes to increased NET formation and elevated level of citrullination. However, as no physical interaction between A20 and PAD4 are found by mass spectrometry, A20 might regulate the expression of PAD4 indirectly and a multistep mechanism might be involved.36 Additionally, citrullination of antimicrobial peptide LL37 is found in cutaneous lupus (LE)/systemic lupus erythematosus (SLE), which may affect the inflammatory properties.37
Compared with other types of PTM, the role of citrullination is not fully elucidated in skin diseases. So far, the identified substrates of PAD include keratin, S100 protein family members and LL37. Whether other proteins such as transcriptional factors could be potentially citrullinated are needed to be clarified in future studies.
Glycosylation
Glycosylation is one of the most common and abundant PTM of proteins. This process is cooperatively regulated by a series of enzymes. Glycosyltransferases, the “writer” of glycosylation, catalyze the transfer of a glycosyl residue from the donor protein to the acceptor protein. Glucosidases, the “eraser” of this process, remove glucose residues from oligosaccharide complex. So far, among various types of glycosylation identified, O-GlcNAcylation, N-glycosylation are the two most common types. So far, approximately 180 glycosyltransferases have been identified and the major enzymes include GNT-V, GNT-III, ST6GAL1 and FUT8.38
It is well known that aberrant glycosylation, impaired localization and expression of glycosyltransferases are strongly related to the onset and progression of various diseases including cancer, inflammatory and autoimmune diseases.39 Therefore, elucidating how glycan structures are altered and identifying the responsible glycosyltransferases and glucosidases are of pivotal issue for understanding disease pathogenesis. FUT8, a member of the glycosyltransferases, is the unique enzyme responsible for generating a core fucose structure on N-glycans. In psoriasis, it was found that FUT8 expression was upregulated in skin tissues, which correlated with disease severity. By mass spectrometry and bioinformatics analysis, EGFR, a receptor upregulated in psoriasis, was identified as the substrate of FUT8 in psoriatic keratinocyte. FUT8 increased the core-fucosylation of EGFR, which subsequently activated keratinocyte proliferation and contributed to the onset of psoriasis.40 Moreover, by assessing the serum level of N-glycans, Zou et al41 identified four N-glycans to be upregulated in the serum of psoriatic patients. However, enlarged size of patients are needed to further validate whether N-glycan profiling could be used in clinical diagnosis and prognosis of psoriasis. Altogether, the above findings suggest the involvement of glycosylation in the pathogenesis of psoriasis.
The involvement of glycosylation in cancer pathology is extensively investigated. One progress is that a series of glycan-based molecules are used as cancer biomarkers, which include AFP, CA125, HE4, CEA and Thyroglobulin.42 A recent review summarizes the specific role of N-glycosylation, especially the glycosyltransferases and their target proteins in skin cancers. Hereby, we update and focus on the role of glycosylation in the pathogenesis of melanomas. Firstly, the aberration of glycosyltransferases were characterized in melanomas.43 Recently, a critical role of FUT8 in promoting melanoma metastasis was uncovered. Glycomic analysis firstly found an elevated core fucosylation level in metastatic melanoma and identified FUT8 as a candidate glycosyltransferase. By using bioinformatic analysis, TGIF2, a transcription factor was identified to be responsible for the upregulation of FUT8 in melanomas. Furthermore, the substrate of FUT8, L1CAM was also found. FUT8 induced the core fucosylation of L1CAM, which reduced its cleavage by proteases and promotes melanoma metastasis.43 GLT8D1, another glycosyltransferase, is reported to be upregulated in melanoma tissues, which showed correlation with clinical features such as histopathological types of melanoma patients.44 These aberrant expressions of glycosyltransferases, which correlated with survival and disease histopathological feathers, might represent as possible diagnostic and prognostic biomarkers of melanoma in the future.
Secondly, novel target proteins of glycosylation were clarified. Among the substrates, STAT5 is a novel identified target of glycosylation, which is an important mediator of EMT process and tumor growth. Scavenger receptor class B type I (SCARB1/SR-BI) is found to be highly expressed in human metastatic melanoma, which contributes to enhanced STAT5 glycosylation, target gene expression and for maintaining EMT phenotypes.45 Moreover, the possible underlying mechanism accounting for glycosylation-induced migration were ongoing investigated. Surprisingly, one study found that different types of glycosylation contributed differentially to melanoma cell migration. Enhanced O-GlcNAcylation exerts a negative role on melanoma cell migration, while N-glycosylation and O-GalNAc glycans positively regulate cell migration. It is suggested that N-glycosylation promotes cell migration by promoting the activity of matrix metalloproteinases MMP2 and MMP9.46
Despite remarkable advances in understanding the involvement of glycosylation in skin diseases, there is a clear need to identify the role of diverse glycosyltransferases in different diseases and to develop new therapeutic targets for the treatment of skin diseases.
Other Types
Despite the aforementioned common types of PTM, other types such as ubiquitination, sumoylation and the newly identified palmitoylation and lysine 2-hydroxyisobutyrylation have also been studied in skin diseases. In 2018,47 our group has summarized the function of ubiquitination in the pathogenesis of psoriasis, so we will not emphasize the role of ubiquitination herein.
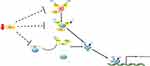
Sumoylation is the covalent conjugation of small ubiquitin-like modifier (SUMO) to target proteins. Like ubiquitination, sumoylation is a reversible process that is catalyzed by E1, E2, E3 enzymes and SUMO-specific peptidases (SENP). In melanoma, the role of sumoylation has already been recognized in the past ten years. In 2007, Ganesan et al48 conducted a study to comprehensively identify SUMO targets in melanoma cells, aiming at interpretation of the interrelationship between SUMO modification, cellular signaling and melanomagenesis. The identified SUMO targets encompass proteins involved in cell growth, signal transduction and transcription. Later, PIASy, a specific SUMO E3 ligase, was found to be responsible for the p53 inactivation in melanoma. PIASy targets p53 for sumoylation, which is responsible for its activation and cell death activity. In melanoma tissues, a decreased expression of PIASy was observed, resulting in the inhibition of p53-mediated cell death in melanoma.49 PIASy can also positively regulate p53 activity via interacting with a DNA polymerase, REV1. Under starvation, PIASy interacts with REV1 and promotes its sumoylation, which enabled its dissociation from p53 and relieved REV1-dependent repression of p53 transactivation (Figure 1).50 In vitiligo which is manifested by asymptomatic white macules resulting from a functional loss of melanocytes in epidermis, the role of sumoylation was recently been identified. Vitiligo lesions display a reduced expression of E2 ubiquitin-conjugating enzyme Ubc9 and an increased expression of SENP1, resulting in an impaired balance between sumoylation and desumoylation. Moreover, knockdown of SUMO1 reduced cell proliferation and led to keratinocyte apoptosis, which might contribute to passive melanocyte death and development of vitiligo.51 Thus, the dynamic nature of sumoylation and its important roles in skin diseases deserves further studies for the development of novel targets for skin therapy.
In psoriasis, the role of some newly identified types of PTM was also studied. Lysine 2-hydroxyisobutyrylation (Khib), a conserved modification, was compared between lesional and nonlesional psoriasis patients. Khib was upregulated in 72 protein and downregulated in 44 proteins. Bioinformatics analysis represented by GO and KEGG pathway suggested an involvement of PI3K-AKT pathway in lysine 2-hydroxyisobutyrylation in psoriasis. Particularly, the most upregulated Khib found in tenasin and downregulated Khib found in S100A9 provide basis for further investigations into the role of Khib in psoriasis.52 Palmitoylation, a reversible PTM which is catalyzed by palmitoyltransferases and depalmitoylating enzymes, is a process that transfers a 16-carbon palmitoyl group onto the protein target. Recently, Zhou et al53 reported the role of a palmitoyltransferase ZDHHC2 in the progress of psoriasis. In IMQ-induced psoriasis mouse models, Zdhhc2 deficiency significantly reduced the pathological grade and the infiltration of pDC by interfering with IRF7 activity to reduce the secretion of pro-inflammatory cytokine IFN-α.53 Thus, the upregulated expression of Zdhhc2 in the epidermis of psoriasis-mouse model provides insights into the role of palmitoylation in the pathogenesis of psoriasis.
Therapeutics Targeting PTM in the Treatment of Skin Diseases
Although a series of studies substantiated the involvement of PTM in inflammatory skin diseases and skin cancers, few studies focused on the therapeutic effect of targeting PTM in skin disease. Herein, we focused on two family members, SIRTs and PARPs, regarding their therapeutic application in skin diseases. Although the study of SIRTs in dermatology is in early stages, the anti-inflammatory role of SIRT1, a member of SIRT family has been well clarified. SIRT1 was reported to deacetylate key transcriptional factors such as NF-κB and AP-1, leading the transcriptional repression of inflammation-related genes.54,55 Regarding the downregulated expression of SIRT1 in psoriasis, SIRT1 activation might represent a promising treatment option for psoriasis patients. SRT2104 is a selective activator of SIRT1 that has been tested in multiple trials. In a randomized, Phase II clinical trial in moderate-to-severe psoriasis patients (n = 40), the effect of SRT2104 was evaluated. Histological assessment showed a significant reduction in epidermal thickness in 35% patients, accompanying with a reduced expression of TNF-α and IL-17 respondergenes and psoriasis-related antimicrobial peptides such as S100A12.56 There are other times, however, where the inhibition of SIRT1 expression might be the desired outcome. In melanoma, SIRT1 overexpression has been previously reported,57 inhibitors of SIRTs have shown potential therapeutic effects in melanomas. Tenovin-1, which inhibits SIRT1, SIRT2 and SIRT3, delays the melanoma growth via induction of p53.58 In a study which used 2 inhibitors of SIRTs, Sirtinol and Ex-527, the anti-proliferative effect of melanoma was also observed.59 However, whether the specific inhibition of one SIRTs or the combined inhibition would achieve better effect in melanoma treatment warrant further investigations.
PARP is activated by repair of DNA damage, which is implicated in drug resistance in tumors and is involved in UV-induced photodamage. PARP1 inhibitors, which have been approved for the therapy of various tumors,60 were also tested for their efficacy in skin diseases. Atopic eczema is a chronic inflammatory skin disease characterized by itchy and generalized skin dryness. An emollient containing nicotinamide, an inhibitor of PARP1 recognized for its anti-inflammatory function, was tested in patients with inflammatory skin diseases including atopic dermatitis, eczema and contact dermatitis. The outcome was very positive and nearly 95% felt an amelioration of their skin condition. The application of nicotinamide also reduced the reliance of patients on topical corticosteroids, suggesting that PARP1 inhibitor might be a potential substitute for the traditional immunomodulators or corticosteroids.61
Altogether, these findings substantiate the potential for SIRTs and PARP inhibitors or activators as disease treatments and preventatives. It is already quite clear that before a drug is used on patients that molecule is subjected to the trial for safety, dosage, etc. There are still many challenges that are needed to be conquered. Firstly, the crosstalk between differential PTM such as acetylation, ubiquitination, PARylation and glycosylation makes this network so complex. Secondly, the wide spectrum of substrates targeting by one specific enzyme are often neglected. Lastly, the current inhibitors and activators are often not specific, which target more than one family members. Regarding these challenges and limits, more specific drugs targeting PTM system should be developed. These drugs might represent as a new generation of therapeutics for the treatment of skin disorders.
Concluding Remarks
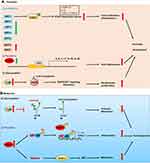
To summarize, we discuss the implication of the major types of PTMs including glycosylation, parylation, acetylation and citrullination in skin diseases. Although many of them have been discovered for many years, only in the last decade the crucial roles of PTMs have attracted considerable attention of researchers in dermatology due to discoveries concerning their enzymology, function and mechanism. Moreover, with the advent of new proteomic techniques, new types of PTMs have shown their importance in various skin diseases, such as carbamylation,62 succinylation,63 etc. Accumulating scientific and clinical evidences support that these PTMs regulate diverse cellular process such as cell differentiation, proliferation, inflammation, metastasis and autophagy. The aberrant expression of enzymes involved in these processes such as glycosyltransferases, PADs, acetyltransferases have been identified in inflammatory, autoimmune skin diseases and skin cancers (Figure 2). However, the substrates of these enzymes, for instance, remain obscure. In citrullination, only keratins, S100 family members and LL37 have been identified as potential substrates. Whether proteins implicated in signaling pathways such as STAT3, NF-κB are substrates of PADs? What is the role of each PAD in specific skin diseases such as psoriasis, atopic dermatitis and skin cancers?
Moreover, the interplay between differential PTMs add to the complexity of illustrating the precise mechanism in skin diseases. What are the dominant PTMs in each specific skin disease? For example, our group has found that ubiquitination of p65 subunit of NF-κB promoted its phosphorylation in psoriasis.64 Furthermore, additional PTMs of p65 are likely to exist, especially acetylation and sumoylation. Do they compete for the binding site of p65 or do they have other modes to function? The answers to these and many other questions will be of great interest to PTMs researchers, immunologists as they could lead to the identification of new therapeutic targets in inflammation-related skin diseases, skin cancers and autoimmune skin diseases.
Acknowledgment
This work was supported by Fundamental Research Funds for the Central University (No. GK202103074, No. GK202103079).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas. All authors gave final approval of the version to be published and have agreed on the journal to which the article has been submitted.
Disclosure
The authors declare no conflicts of interest.
References
1. Chen J, Liu Q, Zeng L, Huang X. Protein acetylation/deacetylation: a potential strategy for fungal infection control. Front Microbiol. 2020;11:2505. doi:10.3389/fmicb.2020.574736
2. Hao L, Park J, Jang H-Y, Bae EJ, Park B-H. Inhibiting protein kinase activity of pyruvate kinase M2 by SIRT2 deacetylase attenuates psoriasis. J Investig Dermatol. 2021;141(2):355–363.e6. doi:10.1016/j.jid.2020.06.024
3. Giblin W, Bringman-Rodenbarger L, Guo AH, et al. The deacylase SIRT5 supports melanoma viability by influencing chromatin dynamics. J Clin Invest. 2021;131(12):e138926. doi:10.1172/JCI138926
4. Fan X, Yan K, Meng Q, et al. Abnormal expression of SIRTs in psoriasis: decreased expression of SIRT 1-5 and increased expression of SIRT 6 and 7. Int J Mol Med. 2019;44(1):157–171. doi:10.3892/ijmm.2019.4173
5. Xu F, Xu J, Xiong X, Deng Y. Salidroside inhibits MAPK, NF-κB, and STAT3 pathways in psoriasis-associated oxidative stress via SIRT1 activation. Redox Rep. 2019;24(1):70–74. doi:10.1080/13510002.2019.1658377
6. Sestito R, Madonna S, Scarponi C, et al. STAT3-dependent effects of IL-22 in human keratinocytes are counterregulated by sirtuin 1 through a direct inhibition of STAT3 acetylation. FASEB J. 2011;25(3):916–927. doi:10.1096/fj.10-172288
7. Xie S, Su Z, Zhang B, et al. SIRT1 activation ameliorates aldara-induced psoriasiform phenotype and histology in mice. J Invest Dermatol. 2015;135(7):1915–1918. doi:10.1038/jid.2015.82
8. Yu X, Li H, Zhu M, et al. Involvement of p53 acetylation in growth suppression of cutaneous T-cell lymphomas induced by HDAC Inhibition. J Investig Dermatol. 2020;140(10):2009–2022.e4. doi:10.1016/j.jid.2019.12.041
9. Rodriguez KM, Cohen MS. Chemical genetic methodologies for identifying protein substrates of PARPs. Trends Biochem Sci. 2021. doi:10.1016/j.tibs.2021.07.002
10. Zhu H, Tang YD, Zhan G, Su C, Zheng C. The critical role of PARPs in regulating innate immune responses. Front Immunol. 2021;12:712556. doi:10.3389/fimmu.2021.712556
11. Bohio AA, Sattout A. c-Abl-mediated tyrosine phosphorylation of PARP1 is crucial for expression of proinflammatory genes. J Immunol. 2019;203(6):1521–1531. doi:10.4049/jimmunol.1801616
12. Kong Q, Li Y, Liang Q, Xie J, Li X. SIRT6-PARP1 is involved in HMGB1 polyADP-ribosylation and acetylation and promotes chemotherapy-induced autophagy in leukemia. Cancer Biol Ther. 2020;21(4):320–331. doi:10.1080/15384047.2019.1702397
13. El-Hamoly T, Hegedűs C, Lakatos P, et al. Activation of poly(ADP-ribose) polymerase-1 delays wound healing by regulating keratinocyte migration and production of inflammatory mediators. Mol Med. 2014;20(1):363–371. doi:10.2119/molmed.2014.00130
14. Kiss B, Szántó M, Hegedűs C, et al. Poly(ADP-ribose) polymerase-1 depletion enhances the severity of inflammation in an imiquimod-induced model of psoriasis. Exp Dermatol. 2020;29(1):79–85. doi:10.1111/exd.14061
15. Angiolilli C, Marut W, van der Kroef M, Chouri E, Reedquist KA, Radstake TRDJ. New insights into the genetics and epigenetics of systemic sclerosis. Nat Rev Rheumatol. 2018;14(11):657–673. doi:10.1038/s41584-018-0099-0
16. Zhang Y, Pötter S, Chen CW, et al. Poly(ADP-ribose) polymerase-1 regulates fibroblast activation in systemic sclerosis. Ann Rheum Dis. 2018;77(5):744–751. doi:10.1136/annrheumdis-2017-212265
17. Kupczyk P, Simiczyjew A, Marczuk J, et al. PARP1 as a marker of an aggressive clinical phenotype in cutaneous melanoma—a clinical and an in vitro study. Cells. 2021;10(2):286. doi:10.3390/cells10020286
18. Donizy P, Wu C-L, Mull J, et al. Up-regulation of PARP1 expression significantly correlated with poor survival in mucosal melanomas. Cells. 2020;9(5):1135. doi:10.3390/cells9051135
19. Fratangelo F, Camerlingo R, Carriero MV, et al. Effect of ABT-888 on the apoptosis, motility and invasiveness of BRAFi-resistant melanoma cells. Int J Oncol. 2018;53(3):1149–1159. doi:10.3892/ijo.2018.4457
20. Amiri KI, Ha HC, Smulson ME, Richmond A. Differential regulation of CXC ligand 1 transcription in melanoma cell lines by poly(ADP-ribose) polymerase-1. Oncogene. 2006;25(59):7714–7722. doi:10.1038/sj.onc.1209751
21. Choi J, Xu M, Makowski MM, et al. A common intronic variant of PARP1 confers melanoma risk and mediates melanocyte growth via regulation of MITF. Nat Genet. 2017;49(9):1326–1335. doi:10.1038/ng.3927
22. Rodríguez MI, Peralta-Leal A, O’Valle F, et al. PARP-1 regulates metastatic melanoma through modulation of vimentin-induced malignant transformation. PLoS Genet. 2013;9(6):e1003531. doi:10.1371/journal.pgen.1003531
23. Cau L, Méchin MC, Simon M. Peptidylarginine deiminases and deiminated proteins at the epidermal barrier. Exp Dermatol. 2018;27(8):852–858. doi:10.1111/exd.13684
24. Witalison EE, Thompson PR, Hofseth LJ. Protein arginine deiminases and associated citrullination: physiological functions and diseases associated with dysregulation. Curr Drug Targets. 2015;16(7):700–710. doi:10.2174/1389450116666150202160954
25. Méchin M-C, Takahara H, Simon M. Deimination and peptidylarginine deiminases in skin physiology and diseases. Int J Mol Sci. 2020;21(2):566. doi:10.3390/ijms21020566
26. Neeli I, Radic M. Opposition between PKC isoforms regulates histone deimination and neutrophil extracellular chromatin release. Front Immunol. 2013;4:38. doi:10.3389/fimmu.2013.00038
27. Dowlatshahi EA, van der Voort EA, Arends LR, Nijsten T. Markers of systemic inflammation in psoriasis: a systematic review and meta-analysis. Br J Dermatol. 2013;169(2):266–282. doi:10.1111/bjd.12355
28. Shelef MA, Sokolove J, Lahey LJ, et al. Peptidylarginine deiminase 4 contributes to tumor necrosis factor α-induced inflammatory arthritis. Arthritis Rheumatol. 2014;66(6):1482–1491. doi:10.1002/art.38393
29. Mastronardi FG, Wood DD, Mei J, et al. Increased citrullination of histone H3 in multiple sclerosis brain and animal models of demyelination: a role for tumor necrosis factor-induced peptidylarginine deiminase 4 translocation. J Neurosci. 2006;26(44):11387–11396. doi:10.1523/JNEUROSCI.3349-06.2006
30. Méchin M-C, Cau L, Galliano M-F, et al. Acefylline activates filaggrin deimination by peptidylarginine deiminases in the upper epidermis. J Dermatol Sci. 2016;81(2):101–106. doi:10.1016/j.jdermsci.2015.11.006
31. Ishida-Yamamoto A, Takahashi H, Iizuka H, Senshu T, Akiyama K, Nomura K. Decreased deiminated keratin K1 in psoriatic hyperproliferative epidermis. J Investig Dermatol. 2000;114(4):701–705. doi:10.1046/j.1523-1747.2000.00936.x
32. Curran AM, Naik P, Giles JT, Darrah E. PAD enzymes in rheumatoid arthritis: pathogenic effectors and autoimmune targets. Nat Rev Rheumatol. 2020;16(6):301–315. doi:10.1038/s41584-020-0409-1
33. Chang X, Han J, Pang L, Zhao Y, Yang Y, Shen Z. Increased PADI4 expression in blood and tissues of patients with malignant tumors. BMC Cancer. 2009;9:40. doi:10.1186/1471-2407-9-40
34. Acharya NK, Nagele EP, Han M, et al. Neuronal PAD4 expression and protein citrullination: possible role in production of autoantibodies associated with neurodegenerative disease. J Autoimmun. 2012;38(4):369–380. doi:10.1016/j.jaut.2012.03.004
35. Ü Basmanav FB, Cau L, Tafazzoli A, et al. Mutations in three genes encoding proteins involved in hair shaft formation cause uncombable hair syndrome. Am J Hum Genet. 2016;99(6):1292–1304. doi:10.1016/j.ajhg.2016.10.004
36. Odqvist L, Jevnikar Z, Riise R, et al. Genetic variations in A20 DUB domain provide a genetic link to citrullination and neutrophil extracellular traps in systemic lupus erythematosus. Ann Rheum Dis. 2019;78(10):1363–1370. doi:10.1136/annrheumdis-2019-215434
37. Lande R, Palazzo R, Hammel P, et al. Generation of monoclonal antibodies specific for native LL37 and citrullinated LL37 that discriminate the two LL37 forms in the skin and circulation of cutaneous/systemic lupus erythematosus and rheumatoid arthritis patients. Antibodies. 2020;9(2):14. doi:10.3390/antib9020014
38. Tang L, Chen X, Zhang X, et al. N-glycosylation in progression of skin cancer. Med Oncol. 2019;36(6):50. doi:10.1007/s12032-019-1270-4
39. Ząbczyńska M, Link-Lenczowski P, Pocheć E. Glycosylation in autoimmune diseases. Adv Exp Med Biol. 2021;1325:205–218. doi:10.1007/978-3-030-70115-4_10
40. Kelel M, Yang R-B, Tsai T-F, et al. FUT8 remodeling of EGFR regulates epidermal keratinocyte proliferation during psoriasis development. J Investig Dermatol. 2021;141(3):512–522. doi:10.1016/j.jid.2020.07.030
41. Zou C, Huang C, Yan L, et al. Serum N-glycan profiling as a diagnostic biomarker for the identification and assessment of psoriasis. J Clin Lab Anal. 2021;35:e23711. doi:10.1002/jcla.23711
42. Kirwan A, Utratna M, O’Dwyer ME, Joshi L, Kilcoyne M. Glycosylation-based serum biomarkers for cancer diagnostics and prognostics. Biomed Res Int. 2015;2015:490531. doi:10.1155/2015/490531
43. Agrawal P, Fontanals-Cirera B, Sokolova E, et al. A systems biology approach identifies FUT8 as a driver of melanoma metastasis. Cancer Cell. 2017;31(6):804–819.e7. doi:10.1016/j.ccell.2017.05.007
44. Hu H, Li Z, Zhou Y, et al. GLT8D1 overexpression as a novel prognostic biomarker in human cutaneous melanoma. Melanoma Res. 2019;29(6):612–620. doi:10.1097/cmr.0000000000000631
45. Kinslechner K, Schörghofer D, Schütz B, et al. Malignant phenotypes in metastatic melanoma are governed by SR-BI and its association with glycosylation and STAT5 activation. Mol Cancer Res. 2018;16(1):135–146. doi:10.1158/1541-7786.MCR-17-0292
46. de Queiroz RM, Oliveira IA, Piva B, et al. Hexosamine biosynthetic pathway and glycosylation regulate cell migration in melanoma cells. Front Oncol. 2019;9:116. doi:10.3389/fonc.2019.00116
47. Yang L, Guo W, Zhang S, Wang G. Ubiquitination-proteasome system: a new player in the pathogenesis of psoriasis and clinical implications. J Dermatol Sci. 2018;89(3):219–225. doi:10.1016/j.jdermsci.2017.12.002
48. Ganesan AK, Kho Y, Kim SC, Chen Y, Zhao Y, White MA. Broad spectrum identification of SUMO substrates in melanoma cells. Proteomics. 2007;7(13):2216–2221. doi:10.1002/pmic.200600971
49. Lakhter AJ, Kanginakudru S, Warren S, Touloukian CE, Boissy RE, Naidu SR. Impaired PIASy-Tip60 signaling weakens activation of p53 in melanoma. Melanoma Res. 2013;23(3):213–217. doi:10.1097/CMR.0b013e328361056d
50. Shim HS, Wei M, Brandhorst S, Longo VD. Starvation promotes REV1 SUMOylation and p53-dependent sensitization of melanoma and breast cancer cells. Cancer Res. 2015;75(6):1056–1067. doi:10.1158/0008-5472.can-14-2249
51. Zhou M, Lin F, Xu W, Jin R, Xu A. Decreased SUMOylation of the retinoblastoma protein in keratinocytes during the pathogenesis of vitiligo. Mol Med Rep. 2018;18(3):3469–3475. doi:10.3892/mmr.2018.9299
52. Ge H, Li B, Chen W, et al. Differential occurrence of lysine 2-hydroxyisobutyrylation in psoriasis skin lesions. J Proteomics. 2019;205:103420. doi:10.1016/j.jprot.2019.103420
53. Zhou B, Yang W, Li W, et al. Zdhhc2 is essential for plasmacytoid dendritic cells mediated inflammatory response in psoriasis. Front Immunol. 2020;11:607442. doi:10.3389/fimmu.2020.607442
54. Sun LN, Zhi Z, Chen LY, et al. SIRT1 suppresses colorectal cancer metastasis by transcriptional repression of miR-15b-5p. Cancer Lett. 2017;409:104–115. doi:10.1016/j.canlet.2017.09.001
55. Wang Y, Huo J, Zhang D. Chemerin/ChemR23 axis triggers an inflammatory response in keratinocytes through ROS-sirt1-NF-κB signaling. J Cell Biochem. 2019;120(4):6459–6470. doi:10.1002/jcb.27936
56. Krueger JG, Suárez-Fariñas M, Cueto I, et al. A randomized, placebo-controlled study of SRT2104, a SIRT1 activator, in patients with moderate to severe psoriasis. PLoS One. 2015;10(11):e0142081. doi:10.1371/journal.pone.0142081
57. Sun T, Jiao L, Wang Y, Yu Y, Ming L. SIRT1 induces epithelial-mesenchymal transition by promoting autophagic degradation of E-cadherin in melanoma cells. Cell Death Dis. 2018;9(2):136. doi:10.1038/s41419-017-0167-4
58. Lain S, Hollick JJ, Campbell J, et al. Discovery, in vivo activity, and mechanism of action of a small-molecule p53 activator. Cancer Cell. 2008;13(5):454–463. doi:10.1016/j.ccr.2008.03.004
59. Wilking MJ, Singh CK, Nihal M, Ndiaye MA, Ahmad N. Sirtuin deacetylases: a new target for melanoma management. Cell Cycle. 2014;13(18):2821–2826. doi:10.4161/15384101.2014.949085
60. Rizvi A, Merlin MA, Shah GM. Poly (ADP-ribose) polymerase (PARP) inhibition in cancer: potential impact in cancer stem cells and therapeutic implications. Eur J Pharmacol. 2021;911:174546. doi:10.1016/j.ejphar.2021.174546
61. Djokic-Gallagher J, Rosher P, Hart V, Walker J. Steroid-sparing effects and acceptability of a new skin gel containing the anti-inflammatory medicinal substance-nicotinamide. Clin Cosmet Investig Dermatol. 2019;12:545–552. doi:10.2147/ccid.s210444
62. Lande R, Pietraforte I, Mennella A, et al. Complementary effects of carbamylated and citrullinated LL37 in autoimmunity and inflammation in systemic lupus erythematosus. Int J Mol Sci. 2021;22(4):1650. doi:10.3390/ijms22041650
63. Nagai T, Saito M, Tanoue Y, Kai N, Suzuki N. Characterization of collagen from sakhalin taimen skin as useful biomass. Food Technol Biotechnol. 2020;58(4):445–454. doi:10.17113/ftb.58.04.20.6734
64. Yang L, Zhang T. Upregulated E3 ligase tripartite motif-containing protein 21 in psoriatic epidermis ubiquitylates nuclear factor-κB p65 subunit and promotes inflammation in keratinocytes. Br J Dermatol. 2021;184(1):111–122. doi:10.1111/bjd.19057
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.