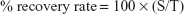Back to Journals » International Journal of Nanomedicine » Volume 14
Efficient recovery and enrichment of infectious rotavirus using separation with antibody-integrated graphite-encapsulated magnetic nanobeads produced by argon/ammonia gas plasma technology
Authors Yamashiro R, Sakudo A, Nagatsu M
Received 24 October 2018
Accepted for publication 8 February 2019
Published 12 March 2019 Volume 2019:14 Pages 1865—1876
DOI https://doi.org/10.2147/IJN.S191784
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Anderson Oliveira Lobo
Risa Yamashiro,1 Akikazu Sakudo,1 Masaaki Nagatsu2
1Laboratory of Biometabolic Chemistry, School of Health Sciences, University of the Ryukyus, Nishihara, Okinawa 903-0215, Japan; 2Research Institute of Electronics, Shizuoka University, Hamamatsu, Shizuoka 432-8561, Japan
Background: Rotavirus is the representative cause of severe acute gastroenteritis in young children. A characteristic feature of rotavirus is low infectious dose and robustness of the virion, suggesting sanitation and hygiene will have little impact. Thus, development of a vaccine should be given priority. Efficient capture of infectious viruses is an important step in generating a vaccine. Previously, antibody-integrated magnetic nanobeads (MNBs) have been developed for virus capture. This study examines the applicability of this method for infectious rotavirus recovery and enrichment.
Materials and methods: Graphite-encapsulated MNBs were treated with radio frequency (RF) excited Ar/NH3 gas mixture plasma to introduce amino groups onto their surfaces. Rotavirus viral protein 7 (VP7) antibody was attached to the surface of MNBs via these amino groups using a coupling agent, N-succinimidyl 3-(2-pyridyldithio)propionate (SPDP). The antibody-integrated MNBs were incubated with rotavirus-infected cell lysate and then separated from the supernatant by applying a magnetic field. After thorough washing, rotavirus was recovered and enrichment analysis done by polymerase chain reaction (PCR), immunochromatography, and an infection analysis using MA104 cells.
Results and discussion: Immunochromatography and PCR indicate that anti-rotavirus antibody-integrated MNPs efficiently capture rotavirus with the capsid protein and viral RNA. The estimated recovery rate was 80.2% by PCR and 90.0% by infection analysis, while the concentrating factor was 7.9-fold by PCR and 6.7-fold by infection analysis. In addition, the absence of non-specific binding to the antibody-integrated MNPs was confirmed using anti-dengue virus antibody-integrated MNBs as a negative control.
Conclusion: These results suggest that this capture procedure is a useful tool for recovery and enrichment of infectious rotavirus. Moreover, when combined with a suitable virus assay this capture procedure can increase the sensitivity of rotavirus detection. Therefore, this capture method is a valuable tool for generating vaccines as well as for developing sensitive detection systems for viruses.
Keywords: rotavirus, antibody-integrated, magnetic beads, nanobeads, nanoparticle
Introduction
Rotavirus, a non-enveloped double-stranded RNA virus, is composed of eleven viral genes encoding six viral proteins (VP), including VP1-4, VP6, and VP7, as well as six non-structural proteins (NSP), including NSP1–6.1,2 Seven rotavirus groups (A to G) are known based on antigenicity of VP6 and sequences of RNA.2 Groups A, B, and C are known to infect human and animal species.3 Group A rotavirus causes severe diarrheal disease and is the predominant cause of acute gastroenteritis in infants and young children worldwide.4–6
Rotavirus is transmitted via the fecal–oral route and multiplies rapidly in cells that line the small intestine causing gastroenteritis.1,2 The feces of an infected individual can contain >10 trillion infectious particles of rotavirus per gram, a small fraction of which is required to infect another person. The common symptom of rotavirus infection is diarrhea following dehydration, which can cause early mortality.7,8 To date, there is no specific cure for rotavirus infection, and patients are only offered symptomatic treatment. The characteristic features of rotavirus include the robustness of the virion and its low infectious dose, with only 100 particles required to cause infection.9 Thus, improved sanitation and hygiene have had little impact on decreasing the incidence of rotavirus infection. A priority of the World Health Organization (WHO) is the development of a vaccine to combat rotavirus infection.10,11 Estimates suggest that vaccination would prevent 2.46 million rotavirus deaths between 2007 and 2025 in countries eligible for vaccine introduction through the Global Alliance for Vaccines and Immunization (GAVI).12 However, to develop a vaccine, an effective collection method of a vaccine source from animals and patient samples or from the environment is needed. For this purpose, an efficient method of concentrating rotavirus would be useful.
There are at least two methods commonly used to concentrate viruses. One is density gradient ultracentrifugation,13 the other is polyethylene glycol (PEG) precipitation.14 Density gradient ultracentrifugation is a time-consuming method that is incompatible with polymerase chain reaction (PCR), a technique used to monitor the concentrating process.15 Although PEG mediated precipitation is a simple method, it is also incompatible with PCR.16,17 Recently, an alternative approach has been studied using a bead-mediated capture method.
Magnetic nanobeads (MNBs) have proved useful in various biomedical applications.18,19 Previously, we have shown that the surface of MNBs can be modified by the introduction of amino groups using radio frequency (RF) ammonia plasma.20 The modified surface of the MNBs can then readily incorporate specific antibodies against a pathogen such as Salmonella,21 dengue virus,22 influenza virus,23 and Escherichia coli.24 Our results show that MNBs decorated with antibodies can facilitate the concentration of these pathogens in environmental samples. However, it is unclear whether MNBs carrying the corresponding antibody on their surface can be used for infectious rotavirus recovery and enrichment.
To examine the applicability of this method for rotavirus recovery and enrichment, we analyzed the binding of rotavirus to anti-rotavirus antibody-integrated MNBs. The efficiency with which rotavirus is concentrated using these beads was assessed by immunochromatography, reverse transcription (RT)-PCR, and real-time RT-PCR as well as an infection analysis.
Materials and methods
Materials
The graphite rod (φ10 mm, 99.99%) was purchased from Nilaco Corp. (Tokyo, Japan), graphite-based adhesive (graphibond-551R) was from Aremco Products, Inc. (Valley Cottage, NY, USA), and Fe2O3 powder (150 μm, 99.99%) was from NewMet Koch (Waltham Abbey, UK). N-succinimidyl 3-(2-pyridyldithio)propionate (SPDP) was purchased from Dojindo Laboratories (Kumamoto, Japan) and dithiothreitol (DTT) was from Wako (Osaka, Japan). The following antibodies were used in this study: mouse rotavirus VP7 monoclonal antibody (MBS319555; MyBiosource, Inc. San Diego, CA, USA), mouse monoclonal anti-dengue virus antibody (clone no NYRDeng2; GeneTex, Inc., Irvine, CA, USA), fluorescein isothiocyanate (FITC)-labeled donkey anti-mouse IgG (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA). Rotavirus Wa strain (VR-2018) and MA104 cells were from American Type Culture Collection (ATCC) (Manassas, VA, USA). Minimum essential medium (MEM) was purchased from Nakalai Tesque (Kyoto, Japan) and fetal bovine serum (FBS) was from JRH Biosciences Inc. (Saint Louis, MO, USA). Penicillin-streptomycin (PS) and trypsin were from Nakalai Tesque. Phosphate buffered saline (PBS) was from Life Technologies (Carlsbad, CA, USA). The adem-Mag SV magnetic device was from Ademtech (Pessac, France). ImmunoCard STAT!® Rotavirus was purchased from Meridian Bioscience Inc. (Cincinnati, OH, USA) and the QIAamp Viral RNA mini kit was from Qiagen (Hilden, Germany). Ex Taq, SYBR Premix Ex Taq II (Tli RNase H plus), T-Vector pMD20 and Thermal Cycler Dice Real Time System were from Takara Bio Inc. (Otsu, Japan). Primers R-20mer and U-19mer were purchased from Novagen (Madison, WI, USA). An ABI373OXL Genetic Analyzer (Applied Biosystems, MA, USA) was used in this study. Bovine serum albumin (BSA) was from Wako, and 4′,6-diamidino-2-phenylindole (DAPI) solution (Cellstain® DAPI) was from Dojindo Laboratories. Fluorescence microscopy was performed using a Biozero BZ-8100 microscope (Keyence, Osaka, Japan). Data analysis was performed with GraphPad Prism 5 software (GraphPad Prism Software Inc., La Jolla, CA, USA).
Plasma-functionalized graphite-encapsulated MNBs and production of antibody-integrated MNBs
The graphite-encapsulated iron compound nanobeads were prepared using an arc discharge method by applying 150–200 A of direct current at ~20 V between an anode and cathode, as described previously.24–26 A graphite rod (φ10 mm, 99.99%; Nilaco Corp.) molded using a graphite-based adhesive (graphibond-551R; Aremco Products, Inc.) with Fe2O3 powder (150 μm, 99.99%; NewMet Koch) was used as the anode. On the opposite side, a high-purity graphite rod was used as the cathode. The resulting graphite-encapsulated iron compound nanobeads (diameter, 20.28±0.84 nm; graphite shell thickness, 3.35±0.23 nm; zeta potential, −9.98~−11.1 mV)26 were then exposed to low-pressure Ar/NH3 gas mixture plasma, produced using an RF power supply at a frequency of 13.56 MHz and a power of 80 W via a matching network.24,27 Initial pre-treatment was performed for 5 minutes using pure Ar gas plasma, followed by 6% NH3 gas added to the Ar plasma post-treatment for 3 minutes to introduce the amino groups. The gas pressure was maintained at 50 Pa throughout pre- and post-treatments. The amino groups on the surface of the MNBs were then reacted with 0.3 μM of the coupling agent, SPDP (Dojindo Laboratories), at pH 7–8. The anti-rotavirus antibody (mouse rotavirus VP7 monoclonal antibody, MBS319555, MyBiosource, Inc.) and the anti-dengue virus antibody (mouse monoclonal antibody; clone no NYRDeng2) were reduced using DTT, leading to breakage of the S–S bonds and generation of S–H groups. The S-H groups on the antibody were then reacted with the SPDP-NH-MNBs, resulting in covalent crosslinking of the antibody onto the surface of the beads. These MNBs were termed antibody-integrated MNBs (Figure 1).
Virus and cells
Rotavirus Wa strain [VR-2018; ATCC] was used as the test virus. MA104 cells (ATCC) were used for infection with rotavirus.
Preparation of the cell lysate
MA104 cells were maintained in MEM supplemented with 10% FBS (JRH Biosciences Inc.). MEM was supplemented with PS by 100 units/mL of penicillin, 100 μg/mL of streptomycin (Nakalai Tesque). Rotavirus was incubated with 20 μg/mL trypsin to activate the virus for 20 minutes. Once the MA104 cells were 80% confluent, the rotavirus was adsorbed onto the cell surface using trypsin for 1 hour. Cell medium was then exchanged with 2 μg/mL trypsin and PS-supplemented MEM medium and cultured at 37°C under 5% CO2 for 6 days. The cultured rotavirus-infected cells were suspended in PBS (Life Technologies), centrifugally washed three times, and freeze-thawed at −80°C. Thereafter, the collected cells were disrupted with a 28 G injection needle and suspended in PBS to prepare a rotavirus-infected cell lysate.
Capture of rotavirus
The capture of rotavirus was performed as follows. Briefly, 10 μL of the antibody-integrated MNBs were washed twice with PBS. A 10 μL sample of rotavirus-infected cell lysate was added to the washed beads with 1 mL PBS, and the mixture was incubated for 5 minutes at room temperature. Tubes containing the mixtures were then placed in a magnetic field for 5 minutes using an Adem-Mag SV magnetic device (Ademtech). Following magnetic separation, the supernatant was removed, and the beads washed three times with PBS. The washed beads were resuspended in PBS and subjected to viral RNA extraction, immunochromatography, or infection analysis. After separation, fractions were obtained using anti-rotavirus antibody-integrated MNBs as follows: 1) bead fraction [10 μL of anti-rotavirus antibody-integrated MNBs after incubation with diluted rotavirus-infected cell lysate (RV-BD)], 2) diluted rotavirus sample before incubation with the beads [10 μL of rotavirus-infected cell lysate (10 μL) diluted with PBS (500 μL)] (BF), 3) supernatant fraction [supernatant after incubation with anti-rotavirus antibody-integrated MNBs and wash (10 μL of supernatant) (RV-SP)], and 4) total sample containing the same quantity of rotavirus as BD (10 μL of rotavirus-infected cell lysate without any treatment) (TL). Anti-dengue virus antibody-integrated MNBs were used as a negative control. The following fractions were obtained: 1) bead fraction [10 μL of anti-dengue virus antibody-integrated MNBs after incubation with diluted rotavirus-infected cell lysate (DV-BD)], 2) supernatant fraction [supernatant after incubation with anti-dengue virus antibody-integrated MNBs and wash (10 μL of supernatant) (DV-SP)].
Immunochromatography
ImmunoCard STAT!® Rotavirus (Meridian Bioscience Inc.) was used to detect rotavirus VP6 in accordance with the manufacturer’s instructions.28
RT-PCR
Viral RNA from samples was extracted with the QIAamp Viral RNA mini kit (Qiagen) according to the manufacturer’s instructions. RNA was extracted by adding lysis buffer prior to removing the beads. RNA was then eluted in 60 μL of nuclease-free water. For the RT-reaction, random primers were added and, after incubation at 25°C for 10 minutes, the RNA was subjected to RT reaction at 65°C for 50 minutes, followed by denaturation of the enzyme at 85°C for 5 minutes. The diluted cDNA was amplified in a reaction mixture containing primers, Ex Taq (Takara Bio Inc.), and Ex Taq buffer by performing 40 cycles of temperature cycling at 94°C for 1 minute, 60°C for 1 minute, and 72°C for 1 minute. PCR was carried out using the following primers for the Rotavirus VP7 gene: forward primer 5′-TGG TGA CTG GAA AGA CTC ATT G-3′; reverse primer 5′-GGA TTA GTT GTT GGA TCT GCT GT-3′. The amplified products were purified and cloned in T-Vector pMD20 (Takara Bio Inc.). DNA sequencing (ABI PRISM3100 Genetic Analyzer, Applied Biosystems) with the R-20mer primer and U-19mer primer (Novagen) was used to verify the product sequence.
Real-time RT-PCR
The viral cDNAs produced by the RT reactions were quantitatively analyzed by real-time PCR using SYBR Premix Ex Taq II (Tli RNase H plus) (Takara Bio Inc.) according to the manufacturer’s instructions (Stratagene). Briefly, the real-time PCR components included SYBR Premix Ex Taq II, and the forward and reverse primers: Forward primer: 5′-GTA CCG TGA AAG TGT GTC CG-3′; Reverse primer: 5′-TCC CAT CAA CGA CAT CCA CT-3′. Real-time PCR was performed using a Thermal Cycler Dice Real Time System (Takara Bio Inc.). The cycling program included initial denaturation at 95°C for 30 seconds followed by 40 cycles of 95°C for 30 seconds and 60°C for 30 seconds. The results were analyzed using Thermal Cycler Dice Real-time System Single software (Takara Bio Inc.). The relative expression ratio of each sample was calculated using a mathematical model based on the amplification efficiency. PCR specificity was verified by dissociation curve analysis of the amplified DNA fragments of step 1 (95°C/15 seconds), step 2 (60°C/30 seconds), and step 3 (95°C/15 seconds). The amplified products were subjected to DNA sequencing after subcloning into Takara T-Vector pMD20 (Takara Bio Inc.), with an ABI373OXL Genetic Analyzer (Applied Biosystems) in order to verify the amplified products.
Infection analysis and indirect fluorescent assay (IFA)
After incubation with the fractions for 1 day, MA104 cells were fixed with 4% paraformaldehyde for 10 minutes and incubated with cold methanol for 5 minutes. The cells were then blocked with 3% BSA for 30 minutes and incubated with anti-rotavirus antibody (Mouse Rotavirus VP7 monoclonal antibody, MBS319555; MyBiosource, Inc.) at 1:50 dilution with PBS for 1 hour at 37°C. Cells were subsequently reacted with FITC-labeled donkey anti-mouse IgG (Jackson ImmunoResearch Laboratories, Inc.) at 1:100 dilution with PBS for 30 minutes at 37°C, and incubated in DAPI solution (Cellstain® DAPI, Dojindo Laboratories) at a final concentration of 1 μg/mL. The cells were visualized under a fluorescence microscope (Biozero BZ-8100; Keyence). The number of rotavirus-infected cells was counted by analyzing 100 cells per field of view in triplicate.
Calculation of recovery rate and concentrating rate
By using data on the number of rotavirus-infected cells and the number of uninfected cells, we calculated the recovery rate and the concentrating rate as follows:
|
|
where T = quantity of genomic RNA or infectivity of total sample containing the same quantity (10 μL) of rotavirus-infected cell lysate as BD (TL); S = quantity of genomic RNA or infectivity of 10 μL of antibody-integrated MNBs after incubation with diluted rotavirus-infected cell lysate (RV-BD or DV-BD); and B = quantity of genomic RNA or infectivity of 10 μL of rotavirus cell lysate with 500 μL of PBS before incubation with the beads (BF).
Statistical analysis
The results are shown as the mean±SD of experiments conducted at least in triplicate. The statistical analysis of significant difference was performed by non-repeated analysis of variance (ANOVA) followed by Bonferroni’s multiple comparison test. Analysis was carried out using GraphPad Prism 5 software (GraphPad Prism Software Inc.).
Results
Detection of viral capsid protein in rotavirus adsorbed onto antibody-integrated MNBs
To investigate the capacity of antibody-integrated MNBs to capture rotavirus, immunochromatography was performed to detect viral capsid protein. Viral capture was performed as described in Figure 2. Immunochromatography showed that the rotavirus VP6 was efficiently concentrated in the anti-rotavirus antibody-integrated bead fraction (RV-BD) compared to the diluted rotavirus sample before incubation with the beads (BF) (Figure 3). Although anti-rotavirus VP7 antibody-integrated MNBs were used for rotavirus capture, rotavirus VP6 was detected in RV-BD. This observation indicates the anti-rotavirus-integrated MNBs recover not only rotavirus VP7, but also other protein components, suggesting that the MNBs can recover the entire rotavirus, including almost all of the viral protein components. In addition, there was no such signal in the anti-dengue virus antibody-integrated bead supernatant fraction (DV-SP), anti-rotavirus antibody-integrated bead supernatant fraction (RV-SP) and anti-dengue virus antibody-integrated bead fraction (DV-BD), suggesting that the capture method for rotavirus using antibody-integrated MNBs was specific.
Detection of viral genomic RNA of rotavirus adsorbed onto antibody-integrated MNBs
Next, RT-PCR was used to detect rotavirus genomic RNA to assess the potential of the beads for capturing rotavirus (Figure 4). RT-PCR analysis gave a single band of 552 bp for VP7 from the anti-rotavirus antibody-integrated bead fraction (RV-BD) and total rotavirus sample containing the same quantity of cell lysate and virus as BD (TL). No signal was detected in the anti-dengue virus antibody-integrated bead supernatant fraction (DV-SP), anti-dengue virus antibody-integrated bead fraction (DV-BD), anti-rotavirus antibody-integrated bead supernatant fraction (RV-SP), or the diluted rotavirus sample before incubation with the beads (BF). The 552 bp band was confirmed to be the viral coat protein VP7 gene of human rotavirus A Wa strain (identical to Genbank accession number FJ423153 100%±0.0%) by DNA sequencing (N=3). Therefore, these results confirm that the anti-rotavirus antibody-integrated bead fraction (RV-BD) includes the rotavirus genomic RNA.
Quantitative analysis of rotavirus genomic RNA in virus adsorbed onto antibody-integrated MNBs
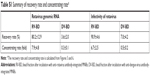
Rotavirus RNA was quantified by real-time RT-PCR in the following aliquots: total rotavirus sample containing the same quantity of cell lysate and virus as BD (TL), diluted rotavirus sample before incubation with the beads (BF), anti-dengue virus antibody-integrated bead supernatant fraction (DV-SP), anti-dengue virus antibody-integrated bead fraction (DV-BD), anti-rotavirus antibody-integrated bead supernatant fraction (RV-SP), and anti-rotavirus antibody-integrated bead fraction (RV-BD) (Figure 5). This analysis showed that the amount of viral RNA corresponding to coat protein VP7 in the anti-rotavirus antibody-integrated bead fraction (RV-BD) was 82.9%±6.9% compared to the TL fraction (104.3%±8.6%). Thus, the recovery rate of RV-BD was 80.2%±12.9% (Table S1). By contrast, the anti-dengue virus antibody-integrated bead fraction (DV-BD) was 3.8%±2.4% compared to the TL fraction, giving a recovery rate for DV-BD of 3.6%±2.0% (Table S1). In addition, the level of rotavirus genomic RNA was below the detectable limit in the anti-dengue virus antibody-integrated bead supernatant fraction (DV-SP), and in the anti-rotavirus antibody-integrated bead supernatant fraction (RV-SP). The real-time PCR product (126 bp) was confirmed to be viral coat protein VP7 of human rotavirus A Wa strain (identical to Genbank accession number FJ423153 100%±0.0%) by DNA sequencing (N=3). Furthermore, as the amount of viral RNA in the diluted rotavirus sample before incubation with the beads (BF) was 12.8%±5.7%, these findings suggest that the anti-rotavirus antibody-integrated MNBs had concentrated the rotavirus by 7.9±4.8-fold (Table S1). By contrast, the anti-dengue virus antibody-integrated MNBs did not concentrate the rotavirus (ie, concentrating rate of 0.3±0.1-fold) (Table S1).
Infectivity of virus adsorbed onto antibody-integrated MNBs
Finally, an infection method using an IFA with anti-rotavirus antibody against rotavirus capsid protein VP7 was performed after incubation of MA104 cells with the fractions for 1 day (Figure S1). Nuclei of all the cells were stained blue with DAPI, while rotavirus-infected cells were stained green with FITC-labeled antibody. A proliferation of rotavirus in MA104 cells was observed in the total rotavirus sample containing the same quantity of cell lysate and virus as BD (TL), and anti-rotavirus antibody-integrated bead fraction (RV-BD). However, the signal was very low in the diluted rotavirus sample before incubation with the beads (BF), the anti-dengue virus antibody-integrated bead supernatant fraction (DV-SP), the anti-dengue virus antibody-integrated bead fraction (DV-BD), and the anti-rotavirus antibody-integrated bead supernatant fraction (RV-SP). To estimate the infectivity of the fractions recovered with the antibody-integrated MNBs, we calculated the ratio of rotavirus-infected cells (Figure 6). The infection rate in the anti-rotavirus antibody-integrated bead fraction (RV-BD) was 90.0%±3.2%, while that in the total rotavirus sample containing the same quantity of cell lysate and virus as BD (TL) was 99.4%±2.1%. By contrast, the infection rate of the anti-dengue virus antibody-integrated bead fraction (DV-BD) was 7.0%±4.3%. In addition, the infection rate in the diluted rotavirus sample before incubation with the beads (BF) was 14.8%±5.2%. Based on these findings, the concentrating rate and recovery rate of the anti-rotavirus antibody-integrated bead fraction (RV-BD) were 6.7±2.5-fold and 90.9%±4.6%, respectively (Table S1). By contrast, the concentrating rate and recovery rate of the anti-dengue virus antibody-integrated bead fraction (DV-BD) were 0.5±0.2-fold and 7.0%±4.2%, respectively (Table S1). Taken together, these results support the notion that infectious rotavirus was efficiently captured and concentrated by the antibody-integrated MNBs without reducing infectivity.
Discussion
In this study, we showed the antibody-integrated MNBs achieved successful enrichment of infectious rotavirus. This may provide a seed vaccine source and enable further development of vaccine candidates using a variety of vaccine production methods. One such method is the Jennerian approach in which an antigenically related rotavirus from an animal host (bovine or simian) is used as the immunogen after cell culture passage to induce protection against human rotavirus.29
Moreover, it should be noted that it is often difficult to differentiate rotavirus gastroenteritis from other causes of gastroenteritis based on clinical symptoms alone.30 Thus, further development of sensitive detection methods for rotavirus would be a valuable aid in confirming the correct diagnosis. The representative detection methods for rotavirus are PCR and antigen-based immunoassay methods (eg, ELISA and immunochromatography).31 ELISA allows quantitative analysis and gives reliable results, but is expensive and time-consuming to perform (3–6 hours). PCR is ~1,000-fold more sensitive than an immunoassay, but has the disadvantage of being both time-consuming (3–6 hours) and a relatively sophisticated technique. Currently, immunochromatography is the most convenient and useful method for obtaining a rapid diagnosis (<30 minutes). However, by coupling these virus detection techniques with an efficient concentrating method using antibody-integrated MNBs, it may be possible to develop a highly sensitive assay procedure for rotavirus.
We previously reported that antibody-integrated MNBs can be used for the efficient capture of Salmonella enterica,21 influenza virus,23 dengue virus,22 and Escherichia coli.24 Therefore, using the amino group-modified MNBs coated with an appropriate antibody, various bacteria and viruses can be captured from liquid samples. This study attempted to examine the capacity of antibody-integrated MNBs to capture rotavirus from liquids. To investigate whether this concentrating method can be combined with virus detection methods, we also performed conventional detection procedures such as immunochromatography, RT-PCR, real-time PCR, and infection analysis.
The present study demonstrated that antibody-integrated MNBs can be used to specifically capture infectious rotavirus. Notably, although VP7 antibody was employed for the capture step, immunochromatography detected rotavirus VP6 protein in the anti-rotavirus antibody-integrated MNB bead fraction (RV-BD) (Figure 3). This result is also supported by PCR analysis. Specifically, PCR analysis detected the transcript of rotavirus VP7 protein in the bead fraction. This finding suggests that the anti-rotavirus antibody-integrated MNBs were able to capture rotavirus containing viral RNA. The PCR results also confirm that our method for capturing rotavirus is compatible with PCR. Furthermore, infection analysis using MA104 cells and IFA indicate that infectious rotavirus can be recovered and enriched using this MNBs capture method.
To evaluate the efficiency of virus capture using the antibody-integrated MNBs, we calculated the recovery rate and concentrating rate using quantitative methods of real-time-PCR as well as an infection analysis. Figure 5 shows the amount of viral RNA for viral coat protein VP7, and Figure 6 shows the infectivity of rotavirus. From the results shown in Figure 5, the recovery rate of the anti-rotavirus antibody-integrated bead fraction (RV-BD) and the anti-dengue virus antibody-integrated bead fraction (DV-BD) were 80.2%±12.9% and 3.6%±2.0%, respectively (Table S1). Figure 6 shows that the recovery rate of the anti-rotavirus antibody-integrated bead fraction (RV-BD) and the anti-dengue virus antibody-integrated bead fraction (DV-BD) were 90.9%±4.6% and 7.0%±4.2%, respectively (Table S1). In addition, from the data of real-time PCR (Figure 5), the concentrating rate of the anti-rotavirus antibody-integrated bead fraction (RV-BD) and the anti-dengue virus antibody-integrated bead fraction (DV-BD) were 7.9±4.8-fold and 0.3±0.1-fold, respectively (Table S1). Likewise, from the data of infection analysis (Figure 6), the concentrating rate of the anti-rotavirus antibody-integrated bead fraction (RV-BD) and the anti-dengue virus antibody-integrated bead fraction (DV-BD) were 6.7±2.5-fold and 0.5±0.2-fold, respectively (Table S1). Taken together, these results show that our capturing method for rotavirus is highly efficient and highly specific.
Although antibody density on the MNB surface may affect their performance, it remains unclear what concentrations of antibody give the best results. Furthermore, the mechanisms by which antibody-integrated MNBs bind to rotavirus remains an open question. It should be noted that MNBs not loaded with antibodies may still contribute to virus binding. Such binding might arise from charged amine groups on the bead surface, which could facilitate their interaction with viruses as shown previously using sulfonated or polyethyleneimine-conjugated MNBs.32,33 Our preliminary experiments indicate that the levels of rotavirus binding to MNBs not loaded with antibodies are much lower than those of antibody-integrated MNBs (Figure S2). However, further studies are needed to elucidate how much antibody binding to the virus contributes to the performance of the beads. To better understand the binding mechanisms, we performed binding experiments of the antibody-integrated MNBs at two temperatures: 4°C and 37°C. No change of rotavirus-binding performance was detected (Figure S3). These observations, combined with further analyses of binding performance under different conditions, may reveal the mechanisms by which the antibody-integrated MNBs bind to rotavirus. In addition, optimizing the parameters for maximizing virus-binding performance by controlling antibody density on the bead surface and/or adjusting the incubation conditions of the antibody-integrated MNBs with the viruses will increase the efficiency of viral recovery and enrichment.
Here, we demonstrate that antibody-integrated MNBs can facilitate the specific capture and concentration of infectious rotavirus from liquid samples. This method is useful for the recovery and enrichment of infectious rotavirus. In addition, the capture method can improve the diagnostic performance of conventional techniques for detecting rotavirus by increasing both sensitivity and specificity. However, the present study used cell lysate as the matrix to analyze the efficacy of rotavirus capture. The efficiency of capture may vary according to the matrix. Thus, further studies are required to analyze a range of different matrices such as feces or other clinical samples from rotavirus-infected animals and patients in place of the cell lysate with varying infectious doses of rotavirus.
Conclusion
Here, we demonstrate that antibody-integrated MNBs are useful for the capture and concentration of infectious rotavirus. The presence of viral protein and viral genome in the captured rotavirus was confirmed by immunochromatography and PCR, respectively. Furthermore, infectivity of the bead fraction was confirmed using an infection analysis. Therefore, this method can contribute to the recovery and enrichment of infectious rotavirus. In addition, by combining the capture procedure with conventional detection methods it will be possible to increase both the sensitivity and specificity of rotavirus assays.
Acknowledgments
This work was supported by Japan Society for the Promotion of Science KAKENHI (Grants-in-Aid for Scientific Research) grant numbers JP16K04997, JP25246029, and JP17H02804. The authors would like to acknowledge Mr Omura (Graduate School of Integrated Science and Technology, Shizuoka University) for assistance in preparing the amino-group functionalized MNBs.
Disclosure
The authors report no conflicts of interest in this work.
References
Esona MD, Gautam R. Rotavirus. Clin Lab Med. 2015;35(2):363–391. | ||
Anderson EJ, Weber SG. Rotavirus infection in adults. Lancet Infect Dis. 2004;4(2):91–99. | ||
Matthijnssens J, Otto PH, Ciarlet M, Desselberger U, van Ranst M, Johne R. VP6-sequence-based cutoff values as a criterion for rotavirus species demarcation. Arch Virol. 2012;157(6):1177–1182. | ||
Matthijnssens J, Ciarlet M, Mcdonald SM, et al. Uniformity of rotavirus strain nomenclature proposed by the rotavirus classification Working Group (RCWG). Arch Virol. 2011;156(8):1397–1413. | ||
Glass RI, Parashar UD, Bresee JS, et al. Rotavirus vaccines: current prospects and future challenges. Lancet. 2006;368(9532):323–332. | ||
Parashar UD, Gibson CJ, Bresee JS, Glass RI. Rotavirus and severe childhood diarrhea. Emerg Infect Dis. 2006;12(2):304–306. | ||
Vesikari T, Matson DO, Dennehy P, et al. Safety and efficacy of a pentavalent Human–Bovine (WC3) reassortant rotavirus vaccine. N Engl J Med. 2006;354(1):23–33. | ||
Ward RL, Bernstein DI, Young EC, Sherwood JR, Knowlton DR, Schiff GM. Human rotavirus studies in volunteers: determination of infectious dose and serological response to infection. J Infect Dis. 1986;154(5):871–880. | ||
Zhan L, Wu WB, Li CM, Huang CZ. Magnetic bead-based sandwich immunoassay for viral pathogen detection by employing gold nanoparticle as carrier. J Anal Test. 2017;1(4):298–305. | ||
Rotavirus. [homepage on the Internet]. Geneva, Switzerland: WHO. Available from: https://www.who.int/immunization/diseases/rotavirus/en/. Accessed October 22, 2018. | ||
Introduction of rotavirus vaccine. [homepage on the Internet]. Geneva, Switzerland: WHO. Available from: http://www.who.int/immunization/monitoring_surveillance/burden/vpd/surveillance_type/sentinel/rotavirus_intro_guidance_who_july31_2013.pdf#search=%27WHO+priority+rotavirus+vaccine%27/. Accessed October 22, 2018. | ||
Atherly DE, Lewis KDC, Tate J, Parashar UD, Rheingans RD. Projected health and economic impact of rotavirus vaccination in GAVI-eligible countries: 2011–2030. Vaccine. 2012;30(Suppl 1):A7–A14. | ||
Arnold M, Patton JT, McDonald SM. Culturing, storage, and quantification of rotaviruses. Curr Protoc Microbiol. 2009;Chapter 15:Unit 15C.3. | ||
Chudzio T, Kasatiya S, Irvine N, Sankar-Mistry P. Rapid screening test for the diagnosis of rotavirus infection. J Clin Microbiol. 1989;27(10):2394–2396. | ||
Roth WK, Weber M, Seifried E. Feasibility and efficacy of routine PCR screening of blood donations for hepatitis C virus, hepatitis B virus, and HIV-1 in a blood-bank setting. Lancet. 1999;353(9150):359–363. | ||
Hamelin C, Lussier G. Concentration of human cytomegalovirus from large volumes of tissue culture fluids. J Gen Virol. 1979;42(1):193–197. | ||
Novotný J, Svobodová J, Ransnäs LA, Kubistová K. A method for the preparation of purified antigens of coxsackievirus B3 from a large volume of cell culture supernatant. Acta Virol. 1992;36(5):483–487. | ||
Sakudo A, Onodera T. Virus capture using anionic polymer-coated magnetic beads (review). Int J Mol Med. 2012;30(1):3–7. | ||
Bergemann C, Müller-Schulte D, Oster J, à Brassard L, Lübbe AS. Magnetic ion-exchange nano- and microparticles for medical, biochemical and molecular biological applications. J Magn Magn Mater. 1999;194(1–3):45–52. | ||
Saraswati TE, Ogino A, Nagatsu M. Plasma-activated immobilization of biomolecules onto graphite-encapsulated magnetic nanoparticles. Carbon. 2012;50(3):1253–1261. | ||
Sakudo A, Chou H, Nagatsu M. Antibody-integrated and functionalized graphite-encapsulated magnetic beads, produced using ammonia gas plasma technology, for capturing Salmonella. Bioorg Med Chem Lett. 2015;25(5):1012–1016. | ||
Sakudo A, Viswan A, Chou H, Sasaki T, Ikuta K, Nagatsu M. Capture of dengue viruses using antibody-integrated graphite-encapsulated magnetic beads produced using gas plasma technology. Mol Med Rep. 2016;14(1):697–704. | ||
Sakudo A, Chou H, Ikuta K, Nagatsu M. Integration of antibody by surface functionalization of graphite-encapsulated magnetic beads using ammonia gas plasma technology for capturing influenza A virus. Bioorg Med Chem Lett. 2015;25(9):1876–1879. | ||
Viswan A, Chou H, Sakudo A, Nagatsu M. Bioconjugation efficiency of plasma-functionalized carbon-encapsulated iron nanoparticles with biotin–avidin system. Biomed Phys Eng Express. 2015;1(4):045104. | ||
Nagatsu M, Yoshida T, Mesko M, et al. Narrow multi-walled carbon nanotubes produced by chemical vapor deposition using graphene layer encapsulated catalytic metal particles. Carbon. 2006;44(15):3336–3341. | ||
Hu R, Furukawa T, Wang X, Nagatsu M. Morphological study of graphite-encapsulated iron composite nanoparticles fabricated by a one-step Arc discharge method. Appl Surf Sci. 2017;416:731–741. | ||
Saraswati TE, Tsumura S, Nagatsu M. High-efficiency plasma surface modification of graphite-encapsulated magnetic nanoparticles using a pulsed particle explosion technique. Jpn J Appl Phys. 2014;53(1):010205. | ||
Meridian Bioscience, Inc. [homepage on the Internet]. Products: ImmunoCard STAT!® Rotavirus. Available from: http://www.meridianbioscience.com/diagnostic-products/rotovirus-and-adenovirus/immunocard/immunocard-stat-rotavirus.aspx/. Accessed October 22, 2018. | ||
Kapikian AZ, Hoshino Y, Chanock RM, Perez-Schael I. Jennerian and modified Jennerian approach to vaccination against rotavirus diarrhea using a quadrivalent rhesus rotavirus (RRV) and human-RRV reassortant vaccine. Arch Virol Suppl. 1996;12:163–175. | ||
Poulton J, Tarlow MJ. Diagnosis of rotavirus gastroenteritis by smell. Arch Dis Child. 1987;62(8):851–852. | ||
Donato C, Cowley D, Kirkwood C. Rotavirus. In: White PA, Netzler NE, Hansman GS. Editors. Foodborne viral pathogens. Boca Raton, FL: CRC Press; 2016:179–218. | ||
Uchida E, Kogi M, Oshizawa T, et al. Optimization of the virus concentration method using polyethyleneimine-conjugated magnetic beads and its application to the detection of human hepatitis A, B and C viruses. J Virol Methods. 2007;143(1):95–103. | ||
Iwata A, Satoh K, Murata M, Hikata M, Hayakawa T, Yamaguchi T. Virus concentration using sulfonated magnetic beads to improve sensitivity in nucleic acid amplification tests. Biol Pharm Bull. 2003;26(8):1065–1069. |
Supplementary materials
  | Table S1 Summary of recovery rate and concentrating rate* |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.