Back to Journals » International Journal of Nanomedicine » Volume 10 » Issue 1
Efficient delivery of NF-κB siRNA to human retinal pigment epithelial cells with hyperbranched cationic polysaccharide derivative-based nanoparticles
Authors Liu Z, Gong H, Zeng R, Liang X, zhang L, Yang L, Lan Y
Received 30 September 2014
Accepted for publication 9 November 2014
Published 7 April 2015 Volume 2015:10(1) Pages 2735—2749
DOI https://doi.org/10.2147/IJN.S75188
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Thomas Webster
Zhenzhen Liu,1,* Haijun Gong,2,* Rui Zeng,2 Xuan Liang,1 Li-Ming Zhang,1 Liqun Yang,1,* Yuqing Lan2,*
1Institute of Polymer Science, School of Chemistry and Chemical Engineering, Key Laboratory of Designed Synthesis and Application of Polymer Material, Key Laboratory for Polymeric Composite and Functional Materials of Ministry of Education, Sun Yat-sen University, Guangzhou, People’s Republic of China; 2Department of Ophthalmology, Guangdong Provincial Key Laboratory of Malignant Tumor Epigenetics and Gene Regulation, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, People’s Republic of China
*These authors contributed equally to this work
Abstract: A hyperbranched cationic polysaccharide derivative-mediated small interfering (si)RNA interference strategy was proposed to inhibit nuclear transcription factor-kappa B (NF-κB) activation in human retinal pigment epithelial (hRPE) cells for the gene therapy of diabetic retinopathy. Two hyperbranched cationic polysaccharide derivatives containing the same amount of cationic residues, but with different branching structures and molecular weights, including 3-(dimethylamino)-1-propylamine-conjugated glycogen (DMAPA-Glyp) and amylopectin (DMAPA-Amp) derivatives, were developed for the efficient delivery of NF-κB siRNA into hRPE cells. The DMAPA-Glyp derivative showed lower toxicity against hRPE cells. Furthermore, the DMAPA-Glyp derivative more readily condensed siRNA and then formed the nanoparticles attributed to its higher branching architecture when compared to the DMAPA-Amp derivative. Both DMAPA-Glyp/siRNA and DMAPA-Amp/siRNA nanoparticles were able to protect siRNA from degradation by nuclease in 25% fetal bovine serum. The particle sizes of the DMAPA-Glyp/siRNA nanoparticles (70–120 nm) were smaller than those of the DMAPA-Amp/siRNA nanoparticles (130–180 nm) due to the higher branching architecture and lower molecular weight of the DMAPA-Glyp derivative. In addition, the zeta potentials of the DMAPA-Glyp/siRNA nanoparticles were higher than those of the DMAPA-Glyp/siRNA nanoparticles. As a result, siRNA was much more efficiently transferred into hRPE cells using the DMAPA-Glyp/siRNA nanoparticles rather than the DMAPA-Amp/siRNA nanoparticles. This led to significantly high levels of suppression on the expression levels of NF-κB p65 messenger RNA and protein in the cells transfected with DMAPA-Glyp/siRNA nanoparticles. This work provides a potential approach to promote hyperbranched polysaccharide derivatives as nonviral siRNA vectors for the inhibition of NF-κB activation in hRPE cells.
Keywords: glycogen, amylopectin, siRNA, nuclear transcription factor-kappa B, human retinal pigment epithelial cells
Introduction
Diabetic retinopathy is a common complication of diabetes that can cause retinal damage eventually leading to blindness, and retinal neovascularization is regarded as one of the most important causes of this complication.1 Clinically, the available therapeutic regimens are laser photocoagulation and blocking neovascularization through multiple intravitreal injections of drugs such as antivascular endothelial growth factor agents, protein kinase C inhibitors, and triamcinolone acetonide.2–5 But these effects are transient and require multiple reinjections. As a result, these therapies are usually associated with several undesirable complications such as vitreous hemorrhage, glaucoma, retinal detachment, and endophthalmitis.2,6
An RNA interference approach, especially using small interfering (si)RNAs, has recently received much attention in treating ocular diseases.7,8 As a therapeutic method to block neovascularization, the siRNA approach has incomparable long-term effects and safety effects. It was reported that nuclear transcription factor-kappa B (NF-kB) played an important role in the angiogenesis of diabetic retinopathy.9–11 On the other hand, human retinal pigment epithelial (hRPE) cells form a monolayer between the neuroretina and the choriocapillaris, which are the essential components of the outer blood retinal barrier that maintain physiological and structural balance within the retina.12 Previous work showed that hRPE cells could synthesize the secreted angiogenic peptide vascular permeability factor/vascular endothelial growth factor,13 and it played a major role in proliferative diabetic retinopathy.14 Therefore, hRPE cells may serve as a potential target for drug delivery and gene transfer leading to diabetic retinopathy.6,12,15 With all the aforementioned research in mind, the decision was taken to investigate the inhibition of NF-kB activation through the siRNA strategy in hRPE cells.
Although siRNA has a small size, siRNA transportation across the cellular membrane is hindered due to its hydrophilicity and negative charge. In addition, siRNA is quickly cleared during in vivo circulation before reaching the target disease site.16 Therefore, successful siRNA-mediated therapy would heavily rely on the development of effective siRNA delivery vectors with high transfection efficiencies and minimal cytotoxicities.16,17 Adeno-associated virus-mediated gene therapy has shown promise in several retinal pigment epithelium (RPE) clinical trials, but adeno-associated virus has a limited payload capacity and potential immunogenicity.18 Nonviral gene delivery vectors, especially cationic polymers (eg, polyethyleneimine, chitosan, and polylysine peptides) and liposomes, have attracted much attention in the RPE gene therapeutic strategy.18 Recent work has established that the architecture of cationic polymers affects the stability, delivery, cytotoxicity, and transfection efficiencies of their complexes with genes.19–22 Compared to linear cationic polymers, hyperbranched cationic polymers exhibit higher levels of gene expression and lower cytotoxicity.19–22 Ahmed et al23 reported that hyperbranched glycopolymers were hemocompatible in vitro and featured less toxicity in response to hyperbranched polymers. Amylopectin and glycogen are naturally hyperbranched polysaccharides with nontoxicity and good biocompatibility, and they are also biodegradable.24–27 Both of them are composed of α-D-(1→4) glucose units of the linear chains, which are interlinked by α-D-(1→6) glycosidic linkage forming the branched structure.24–27 In contrast to amylopectin, the fraction of branching units for glycogen is higher at 8%, whereas the value is approximately 5% for amylopectin.25–27 Additionally, the weight average molecular weight (Mw) of glycogen (106-107 g/mol) is usually smaller than that of amylopectin (106-108 g/mol).24–26 We recently reported that both hyperbranched cationic amylopectin and glycogen derivatives conjugated with 3-(dimethylamino)-1-propylamine (named as DMAPA-Amp and DMAPA-Glyp, respectively; Figure 1) exhibited good blood compatibility and low cytotoxicity, and could effectively deliver plasmid DNA in vitro.28,29 However, whether hyperbranched polysaccharide derivatives can effectively deliver siRNA into hRPE cells is unclear.
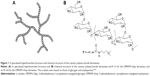  | Figure 1 A speculated hyperbranched structure and chemical structure of the cationic polysaccharide derivatives. |
In this work, the siRNA approach was used to knockdown the NF-kB p65 subunit and inhibited the function of NF-kB, since p65 has been shown to be a key active subunit in NF-kB transcription on hRPE cells.9–11 Nanoparticles based on two hyperbranched cationic polysaccharide derivatives containing the same amount of cationic residues, but different branching structures and molecular weights, including DMAPA-Glyp and DMAPA-Amp derivatives, were developed for the delivery of siRNA into hRPE cells. To optimize one hyperbranched cationic polysaccharide derivative as a potential siRNA delivery vector, the effects of branching structures and molecular weights of these derivatives on cytotoxicity, siRNA delivery efficacy, and NF-kB gene silencing efficiency were investigated.
Materials and methods
Materials
Glycogen (from oysters), amylopectin (from maize), and branched polyethylenimine (bPEI) (Mw =2.5×104 g/mol) were purchased from Sigma-Aldrich Co. (St Louis, MO, USA). The Mw values of glycogen and amylopectin were determined to be 1.5×106 g/mol and 3.1×107 g/mol by static light scattering, respectively. Synthesis of two hyperbranched cationic polysaccharide derivatives (ie, DMAPA-Amp and DMAPA-Glyp) was described in detail in the Supplementary materials. The degree of substitution of DMAPA residues on the polysaccharide, which is defined as the number of DMAPA residues per glucose unit of polysaccharides, was determined to be 2.8 for both polysaccharide derivatives using 1H nuclear magnetic resonance spectroscopy. The weight average molecular weights of the DMAPA-Glyp and DMAPA-Amp were determined to be 4.1×106 g/mol and 8.6×107 g/mol according to the degrees of substitution and the Mw values of the native of glycogen and amylopectin (Table 1).
  | Table 1 Characterization of the cationic hyperbranched polysaccharide derivatives and properties of the polysaccharide derivative/siRNA nanoparticles |
Lipofectamine 2000 (Lip2000), fetal bovine serum (FBS), Dulbecco’s Modified Eagle’s Medium (DMEM), penicillin–streptomycin, trypsin, and Opti-MEM® were purchased from Invitrogen Co. (Carlsbad, CA, USA). Hoechst 33258 was purchased from the Beyotime Institute of Biotechnology (Shanghai, People’s Republic of China), and Cell Counting Kit-8 (CCK-8) from Dojindo Laboratories (Kumamoto, Japan). A siRNA duplex was designed to target human the NF-kB p65 gene based on the public GenBank and applied by Guangzhou RiboBio Co., Ltd. (Guangzhou, People’s Republic of China). It is a 21 bp double-stranded RNA oligos with dTdT 3′ overhangs and has sequences as follows: (sense) 5′-GGACAUAUGAGACCUUCAAdTdT-3′; and (antisense) 5′-UUGAAGGUCUCAUAUGUCCdTdT-3′. A 5′-cy5-labeled nonspecific siRNA duplex (cy5-siRNA) was prepared as a control in the sequence as follows: (sense) 5′-cy5-UUCUCCGAACGUGUCACGUdTdT-3′; and (antisense) 5′-ACGUGACACGUUCGGAGAAdTdT-3′. A nonspecific siRNA duplex was also prepared in the same sequence without cy5 labeling.
Preparation of the hyperbranched polysaccharide derivatives/siRNA complexes
The cationic polysaccharide derivatives were dissolved in RNase-free water (Takara Bio Inc, Kyoto, Japan) at a concentration of 1 mg/mL. Lyophilized siRNA was dissolved in RNase-free water to form a stock solution of 20 μM. A portion of siRNA solution (2.5 μL) was diluted in 200 μL of Opti-MEM® and incubated at room temperature for 5 minutes. Then, the polysaccharide derivative solutions at different polysaccharide derivative/siRNA weight ratios were added to this solution. The resulting mixtures were gently agitated for 10 seconds on a vortex agitator before being incubated at room temperature for 20 minutes prior to use. The nanoparticles containing cy5-siRNA were prepared in the same way in the dark circumstance.
Agarose gel electrophoresis
To assess the condensation ability of the cationic polysaccharide derivatives to siRNA, electrophoresis tests were performed. Ten microliters of the polysaccharide derivative/siRNA nanoparticles with different polysaccharide derivative/siRNA weight ratios in the range of 0.5–20 and naked siRNA were loaded onto 2% agarose gels containing 3 μL of Goldview fluorescence reagent (SBS Genetech Co. Ltd., Beijing, People’s Republic of China) and run with Tris-acetate running buffer at 120 V for 15 minutes. siRNA retardation was then observed and photographed under ultraviolet illumination using an INFINITY 3026 gel image machine (Vilber Lourmat Deutschland GmbH, Eberhardzell, Germany).
Serum stability study
To determine the protective property of the cationic polysaccharides against siRNA degradation, the cationic polysaccharide derivative/siRNA nanoparticles with several weight ratios ranging from 5–20 were incubated with 25% FBS at 37°C for 24 hours, based on the method used in the literature.30 Meanwhile, free siRNA was incubated with 25% FBS as the negative control. Samples were then incubated for 1 hour with excess heparin solution at a heparin/siRNA weight ratio of 5 to ensure complete release of siRNA from the nanoparticles. The samples, as well as an equal weight of intact siRNA without FBS addition, as well as FBS of the same volume, were assessed via agarose gel electrophoresis assay, as described earlier, to examine the integrity of siRNA.
Characterization of the cationic polysaccharide derivative/siRNA nanoparticles
The solutions of polysaccharide derivative/siRNA nanoparticles with different weight ratios (2 mL) were prepared in double-distilled water to a final concentration of 1.0 μg/mL for siRNA, as mentioned previously. Following filtration through a membrane filter (nominal pore size of 0.45 μm), the nanoparticle sizes were measured by dynamic light scattering using ZetaPALS (Brookhaven Instruments Corporation, Holtsville, NY, USA) at 25°C with a 90° scattering angle. The zeta potentials were also evaluated using the same instrument. All of the measurements were performed in triplicate. The morphology of the polysaccharide derivative/siRNA nanoparticles (w/w=20) was observed on an S-4800 scanning electron microscope (HI-9056-0003; Hitachi Ltd., Tokyo, Japan), after the samples were sputter-coated with gold in an E-1045 ion sputter (Hitachi Ltd.).
Cell culture
D407 hRPE cells were provided by the ophthalmic laboratory of the Zhongshan Ophthalmic Center of Sun Yat-sen University (Guangzhou, People’s Republic of China), and this study was approved by the Sun Yat-sen Memorial Hospital’s Ethics Committee (ethics number: 2010-06). The cells were cultured in DMEM containing 10% FBS and 1% antibiotic mixtures of 100 U/mL penicillin G and 100 μg/mL streptomycin (Gibco®; Thermo Fisher Scientific). Before performing a transfection experiment, the cells, which were further identified by immunocytochemical staining, as described in detail in Figure S1, were grown to 30%–40% confluency in a humidified 5% CO2 environment at 37°C.
Cytotoxicity assay
Cytotoxicity of the polysaccharide derivatives and the polysaccharide derivative/siRNA nanoparticles was evaluated using the CCK-8 assay.31 Detailed information was provided in the Supplementary materials.
Evaluation of cell uptake efficiency
To evaluate hRPE cell uptake efficiency of the polysaccharide derivative/cy5-siRNA nanoparticles, flow cytometry assay and laser confocal microscopy were employed. The detailed information was provided in the Supplementary materials.
Evaluation of suppression on NF-kB p65 gene expression
hRPE cells were plated in six-well plates and cultured for 24 hours, following incubation with the polysaccharide derivative/siRNA nanoparticles (w/w=10) for 6 hours, as mentioned previously. Afterward, the medium was changed with fresh DMEM containing 10% FBS. Following an additional 24 hours, 48 hours, and 72 hours of incubation, the medium was removed, and the cells were collected for RNA extraction. Normally, cultured hRPE cells without any treatment were used as the control, and hRPE cells incubated with the Lip2000/nonspecific siRNA complex served as the negative control. The expression levels of NF-kB p65 mRNA and protein were then evaluated in hRPE cells, using semiquantitative and quantitative real-time reverse transcription polymerase chain reaction (PCR) assays, as well as Western blot assay. Details were supplied in the Supplementary materials.
Statistical analysis
Statistical analysis was performed by one-way analysis of variance (SPSS software, version 13.0; IBM Corporation, Armonk, NY, USA). Results were expressed as the mean ± standard deviation, and a value of P<0.05 was considered statistically significant.
Results and discussion
Formation of the cationic polysaccharide derivative/siRNA complexes
Hyperbranched cationic polymers have recently received much attention as a nonviral gene delivery vector because of their high levels of gene expression.19–23 With this in mind, two hyperbranched cationic polysaccharide derivatives containing the same amount of cationic residues, but different branching structures and molecular weights, were developed for delivery of siRNA into hRPE cells to silence NF-kB gene expression (Figure 1 and Table 1).
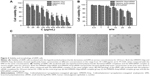
The formation of the cationic polysaccharide derivative/siRNA complexes was examined by agarose gel electrophoresis assay using naked siRNA as a control. As shown in Figure 2A, the migration of siRNA was completely retarded when the DMAPA-Amp derivative/siRNA and the DMAPA-Glyp derivative/siRNA weight ratios exceeded 10 and 5, respectively. The results indicated that both hyperbranched cationic polysaccharide derivatives could condense siRNA to the complexes, which may facilitate cell uptake while providing protection from nuclease degradation. The DMAPA-Glyp derivative formed the complex with siRNA at lower polysaccharide derivative/siRNA weight ratios, attributed to its higher branching architecture, when compared to the DMAPA-Amp derivative. This was in agreement with the literature, which reported that higher branched polymers showed stronger complexation and condensation of nucleic acid.19,21
Zeta potentials and particle sizes
As shown in Figure 3A, the zeta potentials became positive after the polysaccharide derivative/siRNA complexes formed. Moreover, the zeta potentials of the polysaccharide derivative/siRNA complexes increased when increasing their weight ratios in the range of +30–+40 mV, which is amenable to the effective condensation of siRNA. Meanwhile, the volumes of the polysaccharide derivative/siRNA complexes decreased when increasing the weight ratios in the range of 100–200 nm ie, the complexes became progressively smaller during the siRNA condensation process (Figure 3B). The particle size of the DMAPA-Glyp/siRNA complexes was smaller than that of the DMAPA-Amp/siRNA complexes due to the higher branching architecture and lower molecular weight of the DMAPA-Glyp derivative (Table 1).
The morphology of polysaccharide derivative/siRNA complexes was investigated by scanning electron microscopy. As shown in Figure 3C, the complexes at a weight ratio of 20 were observed as nanoparticles with a spherical shape. The size of the DMAPA-Glyp/siRNA nanoparticles (70–120 nm) was significantly smaller than that of the DMAPA-Amp/siRNA nanoparticles (130–180 nm), which is in agreement with the results of the particle size analysis from Figure 3B.
Serum stability
It is considered that, as with cationic polymer/siRNA complexes, high serum stability and low cytotoxicity are essential for successful siRNA delivery both in vitro and, especially, in vivo.30 The serum stability of hyperbranched polysaccharide/siRNA complexes was assessed in 25% FBS (Figure 2B). Naked siRNA was completely degraded by nuclease with 24-hour incubation (Figure 2Bb). However, the migration of siRNA was observed after the polysaccharide derivatives/siRNA complexes were incubated with FBS and treated with heparin solution (Figure 2Bd–i). This demonstrated that both hyperbranched polysaccharides were able to efficiently protect siRNA from degradation by nuclease in serum and release complexed siRNA in the presence of heparin. On the other hand, the weak bands were observed during the siRNA migration process (Figure 2Bb and d–j), which were probably attributed to some components existing in FBS when compared with the weak bands of FBS in Figure 2Bc.
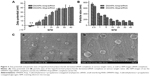
Cytotoxicity
Cytotoxicity of the polysaccharide derivatives was evaluated in hRPE cells using the CCK-8 assay (Figure 4A). Both the DMAPA-Glyp and DMAPA-Amp derivatives showed significantly lower cytotoxicity against hRPE cells when compared to bPEI. In comparison, the DMAPA-Glyp derivative showed lower toxicity against hRPE cells. This may be related to the lower molecular weight of the DMAPA-Glyp derivative and the different biocompatibilities of two native polysaccharides as well. That is, glycogen is from animals and exhibits better biocompatibility for hRPE cells than does amylopectin, which comes from plants.
Although a positive charge was known to facilitate the cell uptake of nanoparticles through a nonspecific electrostatic interaction between the nanoparticles and negatively-charged cell membranes, it was also considered a major cause for cytotoxicity.32 Therefore, it was important to evaluate the cytotoxicity of the polysaccharide derivative/siRNA nanoparticles formed at different w/w ratios. As shown in Figure 4B, the polysaccharide derivative/siRNA nanoparticles exhibited much lower cytotoxicity than did the bPEI/siRNA complexes in the hRPE cells, determined using the CCK-8 assay, owing to lower cytotoxicities of the polysaccharide derivatives. For example, as the w/w ratio reached 20, cells transfected with the polysaccharide derivative/siRNA nanoparticles still retained a relatively high viability of about 80%, while the cell viability reduced to 40% in the cells transfected with the bPEI/siRNA complex. Furthermore, it was found that the cytotoxicity of the DMAPA-Glyp/siRNA nanoparticles were lower than those of the DMAPA-Amp/siRNA nanoparticles due to the lower cytotoxicity of the DMAPA-Glyp derivative.
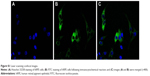
The morphology of hRPE cells incubated with the polysaccharide derivative/siRNA nanoparticles and the bPEI/siRNA complex is shown in Figure 4C. The untreated hRPE cells were used as the control (Figure 4Ca). hRPE cells incubated with both the DMAPA-Glyp/siRNA and DMAPA-Amp/siRNA nanoparticles showed no obvious change (Figure 4Cb and c) in contrast with the control. However, hRPE cells incubated with the bPEI/siRNA complex were destroyed or shrunken, suggesting strong cytotoxicity (Figure 4Cd).
Study on cell uptake
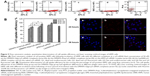
The cell uptake of cationic polymers/gene complexes is assumed to be dependent on their surface properties and particle sizes. It has been reported that cationic polymers/gene complexes with a positive surface charge and with sizes between 50 nm to several hundred nanometers would be suitable for the endocytosis of complexes and efficient gene delivery.33 Based on the aforementioned results, the zeta potentials and particle sizes of both polysaccharide derivative/siRNA nanoparticles were in the range of +30–+40 mV and 100–200 nm, respectively. As expected, these properties enabled the polysaccharide derivative/siRNA nanoparticles to enter the hRPE cells feasibly. The hRPE cell uptake of the polysaccharide/siRNA nanoparticles was evaluated using flow cytometry; this was compared with the bPEI/cy5-siRNA complex and the Lip2000/cy5-siRNA complex as the positive controls, and the naked cy5-siRNA as the negative control (Figures 5A and S2). It was found that there were significantly more fluorescent cells than nonfluorescent cells in Figure 5Aa and b, and in Figure S2A and B, indicating that cy5-siRNA could be effectively transferred into hRPE cells mediated by the DMAPA-Glyp and DMAPA-Amp derivatives. Cell uptake efficiency was then quantitative determined by scoring the percentage of cy5-positive hRPE cells, as shown in Figure 5B. The results indicated that the cell uptake efficiency generally increased when increasing the weight ratios of the polymer/siRNA nanoparticles. Moreover, siRNA was much more efficiently transferred into hRPE cells using the DMAPA-Glyp/siRNA nanoparticles rather than the DMAPA-Amp/siRNA nanoparticles. This is attributed to the smaller sizes and higher zeta potentials of the DMAPA-Glyp/siRNA nanoparticles than those of the DMAPA-Amp/siRNA nanoparticles. It should be noted that the cell uptake efficiency of the DMAPA-Glyp/cy5-siRNA nanoparticles (w/w=20) was close to that of the Lip2000/cy5-siRNA complex and the bPEI/cy5-siRNA complex (w/w=20), implying the high cell uptake efficiency of DMAPA-Glyp/cy5-siRNA nanoparticles.
The cell uptake was further confirmed by the accumulation and spread of red fluorescence of cy5-siRNA in the cytoplasm in the confocal images (Figure 5C), when hRPE cells were incubated for 24 hours and 36 hours, following 6 hours of incubation, with the DMAPA-Glyp/Cy5-siRNA nanoparticles (w/w=10). The spread of red fluorescence of cy5-siRNA in the cytoplasm suggested the endocytosis and endosome escape of the DMAPA-Glyp/siRNA nanoparticles, which could be explained by the good buffer capability of the DMAPA-Glyp derivative.29
Suppression on NF-kB p65 gene expression
NF-kB activation is thought to play an important role in the angiogenesis of diabetic retinopathy, since it leads to the high expression of angiogenesis factors such as cyclooxygenase-2, interleukin-8, and tumor necrosis factor-α, resulting in retinal neovascularization.9–11 Herein, we attempted to block retinal neovascularization through the inhibition of the signal pathway of NF-kB in hRPE cells using a siRNA strategy. siRNA was designed and used to knockdown the NF-kB p65 subunit and inhibited the function of NF-kB, since p65 has been shown to be a key active subunit in NF-kB transcription on hRPE cells.9–11
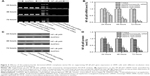
According to the proposed molecular mechanism of the RNA interference approach, cells transfected with effective siRNA can exhibit a reduction in the amount of targeted mRNA and in the amount of protein of NF-kB p65.34 To assess whether polysaccharide derivative/siRNA nanoparticle-mediated gene silencing occurred, the expression levels of NF-kB p65 mRNA and protein were evaluated in hRPE cells. As shown in Figure 6A, the NF-kB p65 mRNA bands progressively weakened when prolonging the incubation times in hRPE cells for both polysaccharide derivative/siRNA groups, as compared to the bands that appeared in the normally cultured control cells and the cells incubated with the Lip2000/nonspecific siRNA complex. The result indicated that the NF-kB p65 mRNA expression was effectively suppressed by both polysaccharide derivative/siRNA nanoparticles. This was further confirmed by quantitative real-time PCR assay, as shown in Figure 6B. It was found that the cells incubated with the DMAPA-Glyp/siRNA nanoparticles showed obviously lower levels of NF-kB p65 mRNA when compared to the cells incubated with the DMAPA-Amp/siRNA nanoparticles.
The determination of suppression at the NF-kB p65 protein level yielded consistent results in the hRPE cells when using Western blot assay, as shown in Figure 6C. hRPE cells incubated with the polysaccharide derivative/siRNA nanoparticles showed extremely lower levels of NF-kB p65 protein when prolonging the incubation times, as compared with the control group and the nonspecific siRNA group. Furthermore, the extremely lower expression level of the NF-kB p65 protein in the DMAPA-Glyp/siRNA nanoparticle group suggested that the DMAPA-Glyp/siRNA nanoparticles showed significantly higher efficacies of suppression on NF-kB p65 than did the DMAPA-Amp/siRNA nanoparticles. For example, the NF-kB p65 protein level in the DMAPA-Glyp/siRNA nanoparticle group was approximately one-third of that in the DMAPA-Amp/siRNA nanoparticle group following 72 hours of incubation in the hRPE cells. This result indicates that the suppression efficacy on gene expression is strongly dependent on cell uptake, which is related to the sizes and charges of the polysaccharide derivative/siRNA nanoparticles.
Conclusion
Two hyperbranched cationic polysaccharide derivatives containing the same amount of cationic residues, but different branching structures and molecular weights, were investigated for the delivery of siRNA into hRPE cells to silence NF-kB gene expression. The DMAPA-Glyp derivative could more efficiently condense siRNA, which was attributed to its higher branching architecture when compared to the DMAPA-Amp derivative. Furthermore, the DMAPA-Glyp derivative showed lower toxicity against hRPE cells. Both hyperbranched cationic polysaccharide derivatives could condense siRNA to form nanoparticles with zeta potentials and particle sizes in the range of +30–+40 mV and 100–200 nm, respectively, which facilitated the protection of siRNA from degradation by nuclease in 25% FBS. The DMAPA-Glyp/siRNA nanoparticles more effectively transferred siRNA to the hRPE cells because of their smaller sizes and higher zeta potentials when compared to the DMAPA-Amp/siRNA nanoparticles. This led to significantly higher levels of suppression on the expression levels of NF-kB p65 mRNA and protein in the cells transfected with DMAPA-Glyp/siRNA nanoparticles.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (21244005, 20974130) and the Science and Technology Planning Project of Guangzhou, Guangdong Province, People’s Republic of China (201300000149).
Disclosure
The authors report no conflicts of interest in this work.
References
Chistiakov DA. Diabetic retinopathy: pathogenic mechanisms and current treatments. Diabetes Metab Syndr. 2011;5(3):165–172. | ||
Porta M, Bandello F. Diabetic retinopathyA clinical update. Diabetologia. 2002;45(12):1617–1634. | ||
Nicholson BP, Schachat AP. A review of clinical trials of anti-VEGF agents for diabetic retinopathy. Graefes Arch Clin Exp Ophthalmol. 2010;248(7):915–930. | ||
Frank RN. Potential new medical therapies for diabetic retinopathy: protein kinase C inhibitors. Am J Ophthalmol. 2002;133(5):693–698. | ||
Jermak CM, Dellacroce JT, Heffez J, Peyman GA. Triamcinolone acetonide in ocular therapeutics. Surv Ophthalmol. 2007;52(5):503–522. | ||
Zhou H, Yang L, Li H, et al. Downregulation of VEGF mRNA expression by triamcinolone acetonide acetate-loaded chitosan derivative nanoparticles in human retinal pigment epithelial cells. Int J Nanomedicine. 2012;7:4649–4660. | ||
Fattal E, Bochot A. Ocular delivery of nucleic acids: antisense oligonucleotides, aptamers and siRNA. Adv Drug Deliv Rev. 2006;58(11):1203–1223. | ||
Oshitari T, Brown D, Roy S. SiRNA strategy against overexpression of extracellular matrix in diabetic retinopathy. Exp Eye Res. 2005;81(1):32–37. | ||
Yoshida A, Yoshida S, Khalil AK, Ishibashi T, Inomata H. Role of NF-kappaB-mediated interleukin-8 expression in intraocular neovascularization. Invest Ophthalmol Vis Sci. 1998;39(7):1097–1106. | ||
Schmedtje JF Jr, Ji YS, DuBois RN, Runge MS. Hypoxia induces cyclooxygenase-2 via the NF-kappaB transcription factor in human vascular endothelial cells. J Biol Chem. 1997;272(1):601–608. | ||
Kroon ME, Koolwijk P, van der Vecht B, van Hinsbergh VW. Hypoxia in combination with FGF-2 induces tube formation by human microvascular endothelial cells in a matrix: involvement of at least two signal transduction pathways. J Cell Sci. 2001;114(Pt 4):825–833. | ||
Song MK, Salam NK, Roufogalis BD, Huang TH. Lycium barbarum (Goji Berry) extracts and its taurine component inhibit PPAR-γ-dependent gene transcription in human retinal pigment epithelial cells: possible implications for diabetic retinopathy treatment. Biochem Pharmacol. 2011;82(9):1209–1218. | ||
Adamis AP, Shima DT, Yeo KT, et al. Synthesis and secretion of vascular permeability factor/vascular endothelial growth factor by human retinal pigment epithelial cells. Biochem Biophys Res Commun. 1993;193(2):631–638. | ||
Hiscott P, Gray R, Grierson I, Gregor Z. Cytokeratin-containing cells in proliferative diabetic retinopathy membranes. Br J Ophthalmol. 1994;78(3):219–222. | ||
Kim YS, Jung DH, Kim NH, Lee YM, Kim JS. Effect of magnolol on TGF-beta1 and fibronectin expression in human retinal pigment epithelial cells under diabetic conditions. Eur J Pharmacol. 2007;562(1–2):12–19. | ||
Lee SJ, Son S, Yhee JY, et al. Structural modification of siRNA for efficient gene silencing. Biotechnol Adv. 2013;31(5):491–503. | ||
Varkouhi AK, Scholte M, Storm G, Haisma HJ. Endosomal escape pathways for delivery of biologicals. J Control Release. 2011;151(3):220–228. | ||
Koirala A, Conley SM, Naash MI. A review of therapeutic prospects of non-viral gene therapy in the retinal pigment epithelium. Biomaterials. 2013;34(29):7158–7167. | ||
Wang R, Zhou L, Zhou Y, et al. Synthesis and gene delivery of poly(amido amine)s with different branched architecture. Biomacromolecules. 2010;11(2):489–495. | ||
Malmo J, Vårum KM, Strand SP. Effect of chitosan chain architecture on gene delivery: comparison of self-branched and linear chitosans. Biomacromolecules. 2011;12(3):721–729. | ||
Fischer D, von Harpe A, Kunath K, Petersen H, Li Y, Kissel T. Copolymers of ethylene imine and N-(2-hydroxyethyl)-ethylene imine as tools to study effects of polymer structure on physicochemical and biological properties of DNA complexes. Bioconjug Chem. 2002;13(5):1124–1133. | ||
Schallon A, Jérôme V, Walther A, Synatschke CV, Müller AHE, Freitag R. Performance of three PDMAEMA-based polycation architectures as gene delivery agents in comparison to linear and branched PEI. Reactive and Functional Polymers. 2010;70(1):1–10. | ||
Ahmed M, Lai BF, Kizhakkedathu JN, Narain R. Hyperbranched glycopolymers for blood biocompatibility. Bioconjug Chem. 2012;23(5):1050–1058. | ||
Putaux JL, Potocki-Véronèse G, Remaud-Simeon M, Buleon A. Alpha-D-glucan-based dendritic nanoparticles prepared by in vitro enzymatic chain extension of glycogen. Biomacromolecules. 2006;7(6):1720–1728. | ||
Rolland-Sabaté A, Mendez-Montealvo MG, Colonna P, Planchot V. Online determination of structural properties and observation of deviations from power law behavior. Biomacromolecules. 2008;9(7):1719–1730. | ||
Rolland-Sabaté A, Colonna P, Mendez-Montealvo MG, Planchot V. Branching features of amylopectins and glycogen determined by asymmetrical flow field flow fractionation coupled with multiangle laser light scattering. Biomacromolecules. 2007;8(8):2520–2532. | ||
de Miranda JA, Cacita N, Okano LT. Evaluation of amylopectin clusters and their interaction with nonionic surfactants. Colloids Surf B Biointerfaces. 2007;60(1):19–27. | ||
Zhou Y, Yang B, Ren X, et al. Hyperbranched cationic amylopectin derivatives for gene delivery. Biomaterials. 2012;33(18):4731–4740. | ||
Liang X, Ren X, Liu Z, et al. An efficient nonviral gene-delivery vector based on hyperbranched cationic glycogen derivatives. Int J Nanomedicine. 2014;9:419–435. | ||
Xiong XB, Uludağ H, Lavasanifar A. Biodegradable amphiphilic poly(ethylene oxide)-block-polyesters with grafted polyamines as supramolecular nanocarriers for efficient siRNA delivery. Biomaterials. 2009;30(2):242–253. | ||
Li XT, Zhang Y, Chen GQ. Nanofibrous polyhydroxyalkanoate matrices as cell growth supporting materials. Biomaterials. 2008;29(27):3720–3728. | ||
Cao N, Cheng D, Zou S, Ai H, Gao J, Shuai X. The synergistic effect of hierarchical assemblies of siRNA and chemotherapeutic drugs co-delivered into hepatic cancer cells. Biomaterials. 2011;32(8):2222–2232. | ||
Liu Y, Reineke TM. Hydroxyl stereochemistry and amine number within poly(glycoamidoamine)s affect intracellular DNA delivery. J Am Chem Soc. 2005;127(9):3004–3015. | ||
Lianxu C, Hongti J, Changlong Y. NF-kappaB p65-specific siRNA inhibits expression of genes of COX-2, NOS-2 and MMP-9 in rat IL-1beta-induced and TNF-alpha-induced chondrocytes. Osteoarthritis Cartilage. 2006;14(4):367–376. |
Supplementary materials
Synthesis of the hyperbranched cationic polysaccharide derivatives
N,N′-carbonyldiimidazole (CDI), 3-(dimethylamino)-1-propylamine (DMAPA), and dimethyl sulfoxide (DMSO) were bought from Aladdin Reagent Co. Ltd. (Shanghai, People’s Republic of China). DMSO was dried for 1 week prior to use by soaking in molecular sieves and calcium hydride. All other chemical reagents were used without further purification. The DMAPA-conjugated amylopectin (DMAPA-Amp) and DMAPA-conjugated glycogen (DMAPA-Glyp) derivatives were synthesized following the methods used in our previous work.1,2 In brief, amylopectin or glycogen (0.1000 g; 0.62 mmol glucose units) was dissolved in 10 mL of water-free DMSO, and was then activated by adding CDI (0.9045 g; 5.58 mmol) and stirred for 1 hour in a nitrogen atmosphere at room temperature. DMAPA (3.8011 g; 37.2 mmol) was added to the amylopectin or glycogen reaction solution. The reaction was allowed to proceed for 24 hours in a nitrogen atmosphere at room temperature. The reaction solution was then dialyzed against the distilled water in a dialysis bag (molecular weight cutoff: 14,000 D) for 3 days, and lyophilized to yield the solid products.
Fourier transform infrared spectroscopy (FTIR) measurement was performed with an FTIR Analyzer (Nicolet/Nexus 670, Thermo Nicolet Corporation, Wisconsin, USA) at a resolution of 4 cm−1 using the KBr method. The assignments of FTIR peaks are listed as follows: 3,325 cm−1 (ν-OH), 2,946 cm−1 (ν-N[CH3]2), 1,709 cm−1, 1,548 cm−1, and 1,258 cm−1 (ν-C=O, δ-NH- and ν-C-N of carbamate groups), 1,462 cm−1 (deformation vibration of –CH2- and –CH3 groups of DMAPA residues), 1,153 cm−1, and 1,036 cm−1 (ν-C-O-C- of polysaccharide).
1H nuclear magnetic resonance (NMR) analysis was carried out on an NMR spectrometer (Mercury Plus 300; Varian Medical Systems, Palo Alto, CA, USA) at 50°C using D2O as a solvent. The signal at δ 4.50 ppm for HDO was used as the internal standard.1,2 The signal assignments are listed as follows: δ=5.00–6.00 (glucose unit, H1); δ=4.50–3.50 (glucose unit, H2–H6); δ=3.27 (–CONH–CH2–); δ=2.55 (–CH2–N<); δ=2.39 (–N[CH3]2), δ=1.83 (–CH2–). The degree of substitution of DMAPA residues on the polysaccharide, which is defined as the number of DMAPA residues per glucose unit of polysaccharides, was determined by the integration of H1 of polysaccharides and protons of DMAPA residues. Degrees of substitution =2.8 for both polysaccharide derivatives.
hRPE cell identification
All human retinal pigment epithelial (hRPE) cells between the third to fifth passages were harvested and cultured on glass coverslips, underwent a process involving a wash three times with phosphate buffered saline (PBS) solution (pH 7.4), fixation with 4% paraformaldehyde for 15 minutes, and a wash three times with PBS solution (pH 7.4) again, before subsequent treatment in 0.1% Triton™ X-100 and 1% bovine serum albumin (50 μL) for 30 minutes. The treated cells were incubated with the primary antibody of anticytokeratin at 4°C overnight, followed by incubation with a second antibody of fluorescein isothiocyanate (FITC)-conjugated goat antimouse antibody immunoglobulin (Ig)G for 30 minutes in darkness; then, the cells were washed three times with PBS solution, the nucleus was stained with Hoechst 33258 (Beyotime Institute of Biotechnology, Shanghai, People’s Republic of China) for 10 minutes, and they were washed three times with PBS solution. Controlled cells were performed using PBS instead of the primary antibody. All hRPE cells were observed with a Zeiss LSM510 confocal microscope (Carl Zeiss Meditec AG, Jena, Germany).
Cytotoxicity assay
hRPE cells (1×104 per well) were seeded in 96-well plates and cultured in a humidified incubator (37°C; 5% CO2) for 24 hours for adherence. The medium was then replaced with 100 μL of fresh medium containing 10 μL of solutions of the polysaccharide derivatives and branched polyethylenimine (bPEI) at different concentrations. In addition, 10 μL of the polysaccharide derivative/small interfering (si)RNA nanoparticles with different weight ratios were added for the cytotoxicity assay, and the amount of siRNA (0.13 μg per well) was constant. The bPEI and the bPEI/siRNA complex were used as the controls. After the cells were incubated for an additional 24 hours, the culture medium was replaced, and then 10 μL of Cell Counting Kit (CCK)-8 reagent was added to each well, following further incubation for 1 hour. The absorbance values of the samples were measured at 450 nm, using a Wellscan MK3 multifunction microplate reader (Thermo Fisher Scientific, Waltham, MA, USA). Cell viability (%) was then calculated according to Equation 1:
|
|
where A450-sample and A450-control were obtained in the presence and absence of the samples, respectively. A450-blank was the absorption of the solution only containing the culture medium and CCK-8 solution without cells. All of the experiments were conducted five times.
The morphology of hRPE cells incubated with the polysaccharide/siRNA nanoparticles and the bPEI/siRNA complex (w/w=10) were observed on a CKX31 inverted phase contrast microscope (Olympus Corporation, Tokyo, Japan).
Flow cytometry analysis
hRPE cells were plated in six-well plates at a density of 4×104 cells/well in 1 mL of Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% fetal bovine serum (FBS), and they were subsequently cultured in a humidified incubator (37°C; 5% CO2) for 24 hours. The solutions of the polysaccharide derivative/cy5-siRNA nanoparticles at different weight ratios (w/w=5, w/w=10, and w/w=20) were prepared and incubated with cells in each well at a cy5-siRNA final concentration of 50 nM. The bPEI/cy5-siRNA and lipofectamine 2000/cy5-siRNA complexes were used as the positive controls. Meanwhile, the naked cy5-siRNA was used as the negative control. The cells were incubated with the complexes in 10% FBS-containing DMEM for 6 hours at 37°C. The media were then changed with fresh DMEM containing 10% FBS. Following further incubation for 24 hours, the cells were washed twice with PBS (pH 7.4) and trypsinized by 0.25% trypsin and suspended in PBS. The cell uptake of cy5-siRNA was detected by an FACSAria™ flow cytometer (BD Biosciences, San Jose, CA, USA). The data were analyzed with CellQuest™ 3.0 software (BD Biosciences). The cell uptake efficiency was then valuated by scoring the percentage of cy5-positive hRPE cells. All of the experiments were conducted in triplicate.
Laser confocal microscopy
hRPE cells (1×104 cells/well) were plated in glass bottom culture dishes for confocal assay and subsequently cultured for 24 hours. The polysaccharide derivative/cy5-siRNA nanoparticles at a weight ratio of 10 were incubated with cells, as mentioned in the experiment of flow cytometry. Following further incubation for 24 hours and 36 hours, the cells were washed three times with PBS and fixed with 4% formaldehyde. They were then stained with Hoechst 33258 (0.1%, w/v) for the nucleus. The intracellular environment of the cy5-siRNA nanoparticles was observed with a Zeiss LSM510 confocal microscope (Carl Zeiss Meditec AG, Jena, Germany).
Semiquantitative RT-PCR assay for the mRNA level of the NF-kB p65 gene
hRPE cells were plated in six-well plates and cultured for 24 hours, following incubation with the polysaccharide derivative/siRNA nanoparticles (w/w=10) for 6 hours, as mentioned previously. Afterward, the medium was changed with fresh DMEM containing 10% FBS. Following an additional 24 hours, 48 hours and 72 hours of incubation, the medium was removed, and the cells were collected for RNA extraction. Normally cultured hRPE cells without any treatment were used as the control, and hRPE cells incubated with nonspecific siRNA served as the negative control.
The messenger (m)RNA level of NF-kB p65 gene was then evaluated using semiquantitative reverse transcription polymerase chain reaction (RT-PCR) assay. Total RNA was isolated from the cells using the RNAiso Plus (Takara Bio Inc., Kyoto, Japan) according to the manufacturer’s manual. Complementary (c)DNA was synthesized by reverse transcription from 1.0 μg of total RNA from each sample using a PrimeScript™ RT reagent Kit (Takara Bio Inc.). The cDNA mixture (1 μL) was used for the semiquantitative RT-PCR amplification mixture (50 μL) containing 25 μL of Premix Ex Taq (Takara Bio Inc.), 1 μL of sense primers (20 μM), and 1 μL of antisense primers (20 μM). Semiquantitative RT-PCR was performed on two genes, NF-kB p65 and GAPDH. The GAPDH gene was measured in each sample as an internal normalization standard. The forward and reverse primers targeting the NF-kB p65 sequence were 5′-TCTCCCTGGTCACCAAGGAC-3′ and 5′-TCATAGAAGCCATCCCGGC-3′, respectively. The forward and reverse primers of GAPDH were 5′-CACCAACTGCTTAGCACCCC-3′ and 5′-TCTTCTGGGTGGCAGTGATG-3′, respectively. For the polymerase chain reaction (PCR), we ran 30 cycles of 98°C for 10 seconds, 68°C for 30 seconds, and 72°C for 30 seconds, and we ran a final extension for 10 minutes at 72°C. All reactions were performed in triplicate. The amplified products were subjected to electrophoresis on 2% agarose gels containing Goldview fluorescence reagent and visualized under an ultraviolet light using ImageJ (National Institutes of Health, Bethesda, MD, USA).
Quantitative real-time RT-PCR assay for the mRNA level of the NF-kB p65 gene
hRPE cells underwent the same siRNA transfection, total RNA extraction, and reverse transcription processes as described previously. Quantitative real-time RT-PCR was performed with LightCycler® 480 SYBR Green I Master reagent and the program was run in a LightCycler® 480 Real-Time PCR System (Hoffman-La Roche Ltd., Basel, Switzerland). Thermal cycling conditions included 5 minutes at 95°C for preincubation, followed by 46 cycles of 10 seconds at 95°C for denaturing, annealing for 15 seconds at 63°C, and extending at 72°C for 10 seconds. Melting-curve analysis was used to confirm the specificity of the amplification reactions. All experiments were carried out in triplicate.
Evaluation of NF-kB p65 gene silencing efficacy by Western blot
hRPE cells underwent the same siRNA transfection process in the experiment of real-time PCR, as was just described. The total protein in the different groups was extracted at 24 hours, 48 hours, and 72 hours after siRNA transfection, and they were then separated using sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes. The membranes were blocked in 5% solution of nonfat milk powder for 1 hour, followed by incubation with the primary antibody of monoclonal rabbit anti-NF-kB p65 (1:600 dilution; Cell Signaling Technology, Danvers, MA, USA) at 4°C overnight. Horseradish peroxidase-conjugated antirabbit IgG (1:8,000 dilution; SouthernBiotech, Birmingham, AL, USA) was used as the secondary antibody, which was further incubated at 37°C for 1 hour. The bands were visualized by the Immobilon Western Chemiluminescent HRP Substrate (EMD Millipore, Billerica, MA, USA). The results were photodocumented and normalized by the GAPDH signal. The percent knockdown of NF-kB p65 was calculated using densitometry from the Western blot image with an ultraviolet light using ImageJ. All experiments were conducted in triplicate.
References
Zhou Y, Yang B, Ren X, et al. Hyperbranched cationic amylopectin derivatives for gene delivery. Biomaterials. 2012;33(18):4731–4740. | ||
Liang X, Ren X, Liu Z, et al. An efficient nonviral gene-delivery vector based on hyperbranched cationic glycogen derivatives. Int J Nanomedicine. 2014;9:419–435. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The
full terms of this license are available at https://www.dovepress.com/terms.php
and incorporate the Creative Commons Attribution
- Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted
without any further permission from Dove Medical Press Limited, provided the work is properly
attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.