Back to Journals » Infection and Drug Resistance » Volume 13
Effect of Short-Term Antimicrobial Therapy on the Tolerance and Antibiotic Resistance of Multidrug-Resistant Staphylococcus capitis
Authors Yu X, Zheng B, Xiao F, Jin Y, Guo L, Xu H, Luo Q, Xiao Y
Received 16 March 2020
Accepted for publication 26 May 2020
Published 30 June 2020 Volume 2020:13 Pages 2017—2026
DOI https://doi.org/10.2147/IDR.S254141
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Xiao Yu,1,* Beiwen Zheng,1,* Feng Xiao,2 Ye Jin,1 Lihua Guo,1 Hao Xu,1 Qixia Luo,1 Yonghong Xiao1
1State Key Laboratory for Diagnosis and Treatment of Infectious Diseases, National Clinical Research Center for Infectious Diseases, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, People’s Republic of China; 2Neurosurgery Department, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yonghong Xiao Email [email protected]
Background: Bacteria undergo adaptive mutation in the host. However, the specific effect of antimicrobial use on bacterial evolution and genome mutations related to bacterial survival within a patient is unclear.
Materials and Methods: Three S. capitis strains were sequentially isolated from cerebrospinal fluid of a clinical inpatient. Antimicrobial susceptibility, growth rate, biofilm formation and whole blood survival of these strains were measured. Relative fitness was calculated. The virulence was examined in the Galleria mellonella model. Whole-genome sequencing and in silico analysis were performed to explore the genetic mechanisms of the changes in antimicrobial resistance phenotype. Hypothetical proteins are cloned, expressed and characterized by detection the susceptibility to gentamycin.
Results: The first isolate was susceptible to rifampin (MIC=0.25 μg/mL), resistant to gentamicin (MIC=16 μg/mL), while the later two isolates were resistant to rifampin (MIC > 64 μg/mL), susceptible to gentamicin (MIC=4 μg/mL). For the latter two strains, compared to the first, frameshift mutation in a hypothetical protein encoding gene and base substitutions (in genes saeR, moaA and rpoB) were discovered. The mutation of rpoB gene caused rifampicin resistance. Mutations in saeR, moaA and hypothetical gene are associated with changes in other biological traits. Amino acid sequence-based structure and function identification of the hypothetical protein indicated that a mutation in the encoding gene might be associated with altered aminoglycoside susceptibility. Growth curve showed that the later two isolates grew faster than the first isolate with a positive fitness advantage of 13.5%, and 14.8%, accordingly. Biofilm form ability and whole blood survival of the derivative mutants were also enhanced. No significant differences of virulence in the G. mellonella model were observed.
Conclusion: We report here for the first time that short-term clinical antibiotic use was associated with resistance mutations, collateral sensitivity, and positive in vivo fitness advantages to S. capitis during infection.
Keywords: resistance, mutations, collateral sensitivity, adaptive enhancement, selective pressure
Introduction
Ever since the first use of antibiotics in the clinic, clinicians have been faced with the emergence of mutations in bacteria that cause antibiotic resistance. Resistance mechanisms allow bacteria to survive in the presence of antimicrobials and are the main reasons for antibiotic treatment failure.1 However, how bacterial genes mutate under the selective pressure of clinical antimicrobial agents, and the effect of these mutations on bacterial resistance and virulence, remain unclear.
The rapid development of high-throughput sequencing technology is providing insight for genomics research, enabling accurate identification of single-nucleotide variations between strains. This approach could reveal genetic evolution of a pathogen within a host, offering new opportunities for exploring the role of genetics and within-host evolution in the outcome of antimicrobial agent-bacterial pathogen interactions. Understanding how a bacterial pathogen evolves during infection of the human host is important for fighting the infection. WGS helps to reveal some important drug-responses of bacteria, such as tolerance, toxicity, and resistance patterns.2 Epidemiological studies of Streptococcus pneumoniae from the last 25 years provide insight into how genomic plasticity within lineages of recombinogenic bacteria permits adaptation to clinical intervention.3 WGS is also used to explore molecular changes in bacterial populations evolving in vivo in relation to long-term chronic infections.4 Such studies revealed that Yersinia pestis harbors different adaptive mutations in different hosts, which increase its ability to infect the host.5 Further, monitoring of resistant bacteria in long-term inpatients led to the identification of recombinant on plasmids.6 Changes in the genome under the selective pressure of antibiotics in vitro are also investigated.3,7 However, few studies have focused on the impact of clinical short-term antimicrobial treatment on the pathogen and collateral sensitivity in S. capitis has not been explored. Mutation analysis of clinical strains isolated at different time points of infection from a patient receiving antibacterial drugs could help explain how pathogens adapt to antibacterial drugs and evolve in the host, and help understand the associated changes in bacterial pathogenicity and other biological traits.
Staphylococcus capitis is a Gram-positive coccus belonging to coagulase-negative Staphylococcus spp. (CoNS), which is frequently found on the human skin and mucosa, and even in the human gut. Although infections caused by this species are rare compared with those caused by Staphylococcus aureus, cases of S. capitis infections are gradually increasing. Recent reports indicate its emergence as a major pathogen causing nosocomial and bloodstream infections.8 Demographic and medical developments that result in increasing numbers of elderly, multimorbid, and immunocompromised patients, and the increasing use of implanted foreign bodies have contributed to the progressively increasing importance of CoNS in healthcare. Furthermore, as for nosocomial pathogens, increasing rates of antibiotic resistance are an even greater problem for CoNS than for S. aureus, from a perspective of therapy, CoNS are challenging because a large proportion of methicillin-resistant strains and increasing numbers of isolates show reduced susceptibility to glycopeptides, limiting the array of available therapeutics.9
In the current study, three strains of S. capitis were isolated from the cerebrospinal fluid of a hospitalized patient. We analyzed these strains by WGS and virulence assays to explore the effect of clinical medication on antimicrobial susceptibility phenotype, virulence and the driving mutations.
Materials and Methods
Ethical Consideration
The current study was conducted in accordance with the Declaration of Helsinki and approved by the ethics committee of The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, China (No:RN.2019.132). Written informed consent was obtained from patient and healthy blood donor.
Bacterial Isolates
Three strains were isolated from the cerebrospinal fluid of a clinical patient with a carotid artery rupture admitted to the First Affiliated Hospital of Zhejiang University. The patient was treated with eight antimicrobial agents such as linezolid, tigecycline, meropenem, polymyxin et al throughout their hospital stay, six antibiotics were introduced after the isolation of the first strain. The isolates were named Sca70935, Sca71187, and Sca71207. The bacteria identification was performed by Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry (MALDI-TOF MS) and WGS. Isolate Sca70935 (the first isolate) was used as a reference strain for the SNPs analysis.
Antimicrobial Susceptibility Testing
Antimicrobial susceptibility of each isolate to penicillin G, oxacillin, gentamicin, clindamycin, erythromycin, quinupristin/dafostine, trimethoprim/sulfamethoxazole, rifampicin, ciprofloxacin, levofloxacin, tigecycline, linezolid, and vancomycin was determined by using the agar dilution method, and interpreted according to the Clinical and Laboratory Standard Institute (CLSI) 2018recommendations.10
Genome Sequencing and Analysis
Genomic DNA was extracted from the three S. capitis isolates by using the Gentra Puregene Yeast/Bact Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. The isolated DNA was sequenced by using the lllumina Novaseq platform. The Trimmomatic software was used to filter low-quality reads. The filtered reads were assembled using the SPAdes Software. Online tools (http://www.genomicepidemiology.org/) were used to identify the acquired antimicrobial resistance genes and the plasmids. For genes whose function could not be annotated by using the RAST Annotation Server, other online tools (https://predictprotein.org/) were used. The command-line PHIGARO software was used for prophage prediction and a web tool CRISPRfinder was used to identify clustered regularly interspaced short palindromic repeats with associated genes (CRISPR/cas). The TMHMM 2.0c program (http://www.cbs.dtu.dk/index.php) was used to predict the presence of transmembrane helices in protein. Structurally similar homologous proteins were identified using the SWISS-MODEL website (https://www.swissmodel.expasy.org) to predict protein function.11 The Snippy pipeline, version 3.0 (https://github.com/tseemann/snippy), was used for read mapping and variant calling.
Growth Assay
The growth curves of these strains were determined by measuring the optical density at a wavelength of 600 nm using an automated growth curve detector (BioTek USA). Briefly, three isolates were cultured overnight in TSB broth and diluted to an optical density at 600 nm (OD600) of 0.01. After dilution 100 times, the bacteria were suspended in MHB medium and grown at 37 °C, with agitation at 200 rpm. The cell density was determined every 0.5 h. The growth curve was plotted using the OD600 value.
Biofilm Assay
The biofilm assay was performed as previously described. Briefly, 1×106 colony-forming units (CFU)/mL were inoculated into TSB broth in polystyrene microtiter 96-well plates and incubated at 37 °C for 24 h. The formed biofilm was stained using crystal violet, the bound dye was eluted with 95% ethanol, and the dye intensity was quantified at OD600. The assay was performed in triplicate; in each replicate assay, the quantification was based on the analysis of eight wells per sample.
Fitness Measurements
The Fitness measurements assay was performed as previously described.12,13 The rifampin-sensitive (RIF-S) isolate Sca70935, the initial isolate, and the two rifampin-resistant (RIF-R) isolates, Sca71187 and Sca71297, isolated subsequently, were diluted to 0.5×106 CFU/mL. Then, equal (10 μL) volumes of Sca70935 and Sca71187 (or Sca71297) cultures were combined, and 20 μL of the mixture was added to 20 mL TSB broth and cultured at 37 °C, with agitation at 200 rpm. After 24 h, 50 μL of the subcultures was inoculated on drug-free MH agar, and 50 μL of the subcultures was inoculated on MH agar containing 8 μg/mL rifampin. The RIF-S and RIF-R colonies were counted the next day, and the adaptive difference was calculated, as follows:
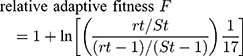
fitness cost C = (1-F)×100%
where rt is the number of resistant colonies and St is the number of sensitive colonies.
Survival of Bacteria in Whole Blood
The survival of bacteria assay was performed as previously described.14 Cultures of the RIF-S and RIF-R S. capitis strains in the logarithmic growth phase (OD600=0.5) were diluted to 2×106 CFU/mL with 0.9% saline; 50 μL of each culture was mixed with 150 μL of the heparinized whole blood from a healthy volunteer in a 1.5 mL Eppendorf tube, and incubated at 37 °C with agitation at 200 rpm. After 0, 60, 120, and 180 min, 10 μL of the mixture was withdrawn, combined with 90 μL of MH broth, and plated on MH agar. Bacterial colonies were counted after overnight incubation, each strain was assayed three times.
Infection of Galleria mellonella Larvae
The virulence of S. capitis isolates was determined by infecting G. mellonella larvae, as described previously.15 Briefly, overnight cultures of S. capitis were washed with phosphate-buffered saline (PBS) and the cell concentration was adjusted with PBS to 1×106 CFU/mL. Each experimental group contained randomly picked 20 G. mellonella larvae weighing 250 mg. Each G. mellonella larva was injected with 20 μL of bacterial suspension; the negative control group larvae were injected with 20 μL of sterile PBS solution. The injected G. mellonella larvae were placed in an incubator at 37 °C and observed every 12 h. The larval survival rate was recorded over 72 h. All experiments were performed in triplicate.
Cloning, Expression and Function Identification of the Hypothetical Protein
Genomic DNA of Sca70935 was extracted by using the Gentra Puregene Yeast/Bact Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. The hypothetical protein encoding gene Anthp1 was amplified using manually designed sequence specific primers (F-GAAGGAGATATACATATGAAACTTGAAGCACAAAAACCA
R-GTGCGGCCGCAAGCTTTTATGATATTTTTCTATTATTTATTACCGCTTC). pET32a vector were digested with Xho I and Nde I restriction enzymes. The PCR amplification was In-Fusion cloned into the digested pET 32a expression vector containing C terminal poly-histidine tag. The clone was further transformed into chemically competent E. coli BL21 (DE3) cells compatible to the expression vector and the integrity of the clone was verified by sequencing. Single colony were grown at 37°C in 5 mL of LB media containing 100 μg/mL ampicillin for 12h. After incubation, cultures was diluted by 1:100 to fresh 50mL LB media containing 1mM isopropyl-1-thio-β-Dgalactopyranoside (IPTG) for expression 3 hours at 37°C at 200 rpm.The fusion protein was observed using SDS-PAGE and characterized by detecting the susceptibility of E. coli BL21 (DE3) to gentamicin in disk diffusion assay.
Accession Numbers
The sequences of Sca70935, Sca71187, and Sca71207have been deposited in GenBank under the accession numbers RYDS00000000, RYDR00000000 and RYDQ00000000.
Results
Basic Clinical Status of the Patient
On January 23, 2019, the patient was transferred to the First Affiliated Hospital of Zhejiang University after an emergency surgery in another hospital because of a rupture of the internal carotid aneurysm. In February 26, Sca70935 was isolated from the patient’s cerebrospinal fluid. The Sca71187 and Sca71207 were obtained on March 5 and March 10, respectively. The patient was treated with eight antimicrobial agents throughout their hospital stay, six antibiotics were introduced after the isolation of the first strain. The medication regime is shown in Figure 1. The patient was discharged on April 3.
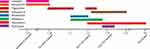 |
Figure 1 Overview of the antibacterial regimen and strain isolation. |
Antimicrobial Susceptibility Profiling
The three isolates were resistant to oxacillin, penicillin G, erythromycin, clindamycin, levofloxacin, moxifloxacin, ciprofloxacin, and linezolid, but were sensitive to sulfamethoxazole, tigecycline, tetracycline, nitrofurantoin, vancomycin, and teicoplanin. Strain Sca70935 was resistant to gentamicin and quinupristin/dalfopristin but sensitive to rifampicin, while strains Sca71187 and Sca71207 were sensitive to gentamicin, intermediate to quinupristin/dalofopine, and resistant to rifampicin. The drug susceptibility data for the three isolates are shown in Table 1.
 |
Table 1 Antimicrobial Susceptibility of the S. capitis Isolates |
Genomic Characteristics of Three Multidrug Resistant Strains
The sizes of the assembled draft genomes of the three isolates were similar, 2.52 Mbp on average. The mean number of contigs in each draft genome was 79. The average N50 value was 22,502 bp and the mean %GC content was 32.82%. The three isolates harbored an identical resistome composed of ten genes, namely, blaZ and mecA for β-lactam resistance; aac(6′)-Ie and ant(4′)-Ib for aminoglycoside resistance; qacA, norA, and mgrA for quinolone resistance; ermA for erythromycin resistance; dfrC for trimethoprim/sulfamethoxazole resistance; and cfrA for linezolid resistance. The genotypes partly explain the resistance profiles (Table 1). The S. capitis strains carry 3 plasmids, a Siphoviridea prophage PHAGE_Staphy_StB20_NC_019915 and no CRISPR/Cas elements in the genome, there is no loss of a plasmid during the treatment.
Mutation Analysis and Function of the Mutated Genes
Compared with the Sca70935 strain, four mutations were identified in Sca71187 and Sca71207 (the two strains harbored the same mutations). Three of these mutations were base substitutions and one was a base deletion, resulting in three missense mutations and one frameshift mutation, respectively (details are shown in Table 2). The base substitutions in rpoB gene and saeR gene were conserved substitutions, while in moaA gene, they were not.
 |
Table 2 Summary of the Protein Variants in the Sca71187 and Sca71207 Strains |
The saeR gene encodes a 228 amino acid polypeptide with an N-terminal regulatory domain and a potential C-terminal DNA-binding domain. The SaeR protein is a member of the two-component regulatory system SaeR/SaeS and is involved in the regulation of expression of virulence genes. It regulates transcription by recognizing a specific DNA sequence near the promoter sequence of the target gene. The SaeR/SaeS system activates the expression of foreign genes and cell wall-associated genes involved in host cell adhesion and invasion, inhibiting the production of type 5 capsular peptidoglycan.16
The second mutated gene, moaA, encodes a protein that catalyzes the cyclization of GTP to (8S)-3′,8-cyclo-7,8-dihydroguanosine 5′-triphosphate, which plays a role in the biosynthesis of molybdenum protein. The molybdenum protein is involved in the biosynthesis of cofactors, which activate and accelerate enzymatic reactions.17 The specific involvement of MoaA in the function of specific S. capitis enzymes is not clear.18
The third mutated gene, rpoB, encodes the β-subunit of the bacterial RNA polymerase, whose main function is to transcribe DNA into RNA. In staphylococci, the rpoB gene contains a rifampicin resistance-determining region; mutations in that region result in resistance to rifampicin.19,20 The mutation in strains Sca71187 and Sca71207 occurs in a cluster I region of the drug resistance-determining region.
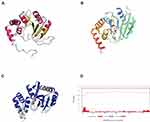
One identified gene encodes a hypothetical protein named Anthp1. We predicted the 3D-structure of the protein based on its amino acid sequence (Figure 2A) and identified protein with 75% sequence coverage in the SWISS-MODEL database (Figure 2B and C). The reference protein (Figure 2B) is an aminoglycosyltransferase involved in the resistance to gentamicin. According to the domain analysis, the encoded hypothetical protein is mainly extracellular (Figure 2D).21 Therefore, we propose that the hypothetical protein may underpin gentamicin resistance in the Sca70935 isolate because, in strains Sca71187 and Sca71207, a 13 C-terminal amino acid stretch, RIEEAVINRKIS, is replaced by NVIKKR (early termination of translation) as a result of a mutation.
Strain Growth and Biofilm Formation
Compared with the initial isolate Sca70935, the growth rate of the other two isolates was increased (p<0.05, independent t-test). There were no significant differences between Sca71187 and Sca71207. After 6 h of growth, the growth rate of the mutant strain significantly increased and continued until plateauing, and the number of the mutant strains in plateau phase was greater than the number of Sca70935 strains (Figure 3A). As shown in Figure 3B, the biofilm-forming ability of the two strains was significantly higher than that of the reference isolate (p<0.001, independent t-test).
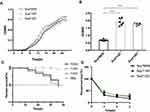 |
Figure 3 (A) Growth curve. (B) Biofilm formation. (C) In vivo infection of G. mellonella larvae. (D) survival of bacteria in whole blood *** indicated (p<0.001). |
The Fitness Cost of Mutations, Bacterial Infection of G. mellonella and Survival of Bacteria in Whole Blood
The relative positive fitness advantage was 13.5% for Sca71187 and 14.8% for Sca71207. Thus, the later derivative mutants all bore an adaptive advantage compared with the parental strain Sca70935. As shown in Figure 3C, G. mellonella died about 50% within 72 h following infection. Kaplan–Meier survival curve analysis showed that the mortality of G. mellonella was similar when infected with the three strains. No significant differences in virulence in the G. mellonella model were observed. The results of the whole blood survival assay are shown in Figure 3D. The whole blood survival of strains Sca71187 and Sca71207 was significantly higher than that of the Sca70935 isolate (p < 0.05).
Effect of Hypothetical Protein in BL21 Strains on Their Susceptibility to Gentamicin
An E. coli strain expressed the previously described hypothetical protein anthp1 (BL21-pet32a-anthp1) were successful constructed to investigate the effect of hypothetical protein on gentamicin resistance. As shown in the result of SDS-PAGE (Figure S1), the protein is clearly expressed. The disk diffusion method showed that the minimum inhibitory zone diameter of gentamicin for strains which express and not express the protein were same 18mm.
Discussion
A wide range of broad-spectrum antibacterial drugs, immunosuppressive agents, and chemotherapeutic drugs is typically used in clinical practice, and various invasive surgeries are widely practiced. This results in an increasing isolation rate of CoNS and the bacterial antibiotic resistance.22 CoNS was once not considered to be pathogenic. However, recent clinical and laboratory tests have confirmed that the majority of normal microbiota are conditional pathogens. Consequently, CoNS has become one of the important pathogens causing hospital bloodstream infections that cannot be easily explained as contamination by opportunistic pathogens.23 According to recent reports, S. capitis is now a major pathogen causing nosocomial and bloodstream infections, meningitis, prosthetic valve endocarditis, and late-onset sepsis. That is mainly because of its ability to produce slimy biofilms, enabling it to adhere to medical devices, such as prosthetic valves and catheters. Hence, the bacterium is difficult to control or clear by the immune response.24 During a clinical treatment, infections caused by S. capitis should be monitored. In particular, the incidence of oxacillin-resistant CoNS is high, in some cases exceeding 60%.25–27
These S. capitis strains isolated in the current study were resistant to methicillin. Of note, these strains were also resistant to linezolid. Linezolid is the first oxazolidinone antibiotic to be approved for clinical use. It is currently used to treat methicillin-resistant S. aureus (MRSA), vancomycin-resistant enterococci (VE), and other important multidrug resistant pathogens. The acquisition of cfr resistance genes and mutations in the 23S rRNA gene constitute the main mechanisms of resistance to these antimicrobials.28
Antimicrobial susceptibility profiling revealed that strains Sca71187 and Sca71207 have become resistant to rifampicin during antimicrobial treatment. According to the clinical data, rifampicin was introduced only after the Sca70935 strain was isolated and confirmed sensitive to rifampicin. The mutation of the rpoB gene was first identified in strain Sca71187 after 5 days of treatment. The point substitution is located in the rifampicin resistance-determining region of the gene. This indicated that the use of rifampicin was associated with rifampicin resistance in S. capitis in a short day, suggesting that the ability of this drug to induce resistance should be monitored in the clinical setting. That is similar to the findings of O’Neill, who reported a high mutation frequency of rifampicin-resistance genes in S. aureus, approximately 10–6–10,–8 which is easily drug-inducible.29
At the same time, susceptibility profiling revealed that during antimicrobial treatment, the gentamicin phenotype of Sca71187 and Sca71207 changed from resistance to susceptibility, and quinupristin/dalofopine resistance changed to intermediate. Collateral sensitivity was first proposed by Szybalski and Bryson in the 1950s, but research on the inducing conditions and molecular mechanisms of this phenomenon has been stagnant in the last few decades.7 Recently, the development of drug resistance and collateral susceptibility have been studied in bacteria exposed to antibiotics via adaptive laboratory evolution, and the molecular mechanisms of collateral synergistic susceptibility were preliminarily explored by WGS. Nevertheless, the mechanism of collateral susceptibility remains unclear.30 Although the three strains harbor genes for two aminoglycoside modifying enzymes, aac (6′)-Ie and ant (4′)-Ib, it has been shown that these two enzymes cause resistance to tobramycin, not gentamicin.31,32 Genome analysis performed in the current study identified another mutated gene in strains Sca71187 and Sca71207. Based on the predicted structure and function of the encoded protein, we speculated that the gene might be involved in gentamicin resistance in strain Sca70935. Although the expression of this protein in E. coli do not decreased the susceptibility to gentamicin, it may play a more important role in S. capitis. Furthermore, structure prediction shows mutation of the encoding gene altered the structure of the C-terminal binding domain of the protein, rendering it unable to bind gentamicin for further modification, and re-sensitizing the bacterium to gentamicin. Alteration of a strain’s sensitivity to rifampicin that alters the sensitivity to other antimicrobial agents might be caused by collateral sensitivity. This effect is widespread during bacterial resistance evolution.33 The mechanism of altered sensitivity to quinupristin/dalofopine observed in the current study is unclear and further research is needed. The re-sensitization of bacteria to gentamicin suggests that clinicians may develop an alternative strategy based on collateral sensitivity to treat bacterial infections, which may inhibit the rise of drug-resistant mutants and delay the development of drug resistance.34–36
SNP analysis of the Sca71187 and Sca71207 genomes revealed mutation in the saeR gene. The saeR gene encodes a response regulator protein, an important element of the SaeRS two-component system. The SaeS sensor detects an environmental signal, following which SaeR binds the target molecule through the C-terminal effector domain and regulates gene transcription. The two-component system is involved in host cell adhesion and invasion, and the expression of genes encoding extracellular proteins and cell wall-associated proteins.37 The experiments performed in the current study demonstrated the increased growth rate and biofilm formation of the mutated isolates, indicating enhanced ability to survive within the host. Therefore, we speculate that an N-terminal mutation lead to more (or less) interaction of SaeR to SaeS and more (or less) expression of genes that are part of the SaeRS regulon. Moreover, these effects could be direct or indirect.
Rifampicin resistance and other changes in the genomes was not associated with an adaptive fitness cost but, instead, promoted bacterial growth rate, biofilm formation, and an ability to survive in the whole blood. This is consistent with other reports that rifampicin-induced resistance is not associated with an adaptive fitness cost.19 This suggests that the effect of a drug on bacterial fitness cost should be considered when using the drug. Further research is needed to investigate the effect of the mutation of the moaA gene on the physiology of bacteria. Mutations of bacteria within the host are key factors that determine the pathogen’s adaptability to the host’s immune system and drug treatment. Understanding the effect of in vivo mutations of bacterial pathogens will facilitate subsequent treatment and prognosis.
Although in the current study we only investigated the mutation of a single strain after antimicrobial treatment, the occurrence of the same mutations in two subsequent isolates indicates the consistency of these mutations. However, more strains should be analyzed in the future.
Conclusion
We reported here for the first time that the adaptive mutations of S. capitis in the cerebrospinal fluid of a clinical patient elicited by antibiotics result in rifampicin resistance, enhanced biofilm formation, and collateral susceptibility with fitness advantage. We preliminary explored the correlation between antimicrobial resistance and changes in protein structure caused by genetic mutations. The results of the current study will help to elucidate the mechanism of the adaptive evolution of S. capitis in response to the selective pressure of antibiotics in the host, to improve the understanding of infection caused by such strains and to devise measures to prevent the emergence of drug resistance based on collateral sensitivity.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (grant number 2017YFC1200203) and the Mega-projects of Science Research of China (grant number 2018ZX10733402-004).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74(3):417–433. doi:10.1128/MMBR.00016-10
2. Harris SR, Clarke IN, Seth-Smith HM, et al. Whole-genome analysis of diverse Chlamydia trachomatis strains identifies phylogenetic relationships masked by current clinical typing. Nat Genet. 2012;44(4):413–419, S411. doi:10.1038/ng.2214
3. Choe D, Lee JH, Yoo M, et al. Adaptive laboratory evolution of a genome-reduced Escherichia coli. Nat Commun. 2019;10(1):935. doi:10.1038/s41467-019-08888-6
4. Marvig RL, Johansen HK, Molin S, Jelsbak L. Genome analysis of a transmissible lineage of pseudomonas aeruginosa reveals pathoadaptive mutations and distinct evolutionary paths of hypermutators. PLoS Genet. 2013;9(9):e1003741. doi:10.1371/journal.pgen.1003741
5. Morelli G, Song Y, Mazzoni CJ, et al. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat Genet. 2010;42(12):1140–1143. doi:10.1038/ng.705
6. Conlan S, Park M, Deming C, et al. Plasmid dynamics in KPC-positive Klebsiella pneumoniae during long-term patient colonization. mBio. 2016;7(3):3. doi:10.1128/mBio.00742-16
7. Lazar V, Pal Singh G, Spohn R, et al. Bacterial evolution of antibiotic hypersensitivity. Mol Syst Biol. 2013;9(1):700. doi:10.1038/msb.2013.57
8. Butin M, Martins-Simoes P, Rasigade JP, Picaud JC, Laurent F. Worldwide endemicity of a multidrug-resistant staphylococcus capitis clone involved in neonatal sepsis. Emerg Infect Dis. 2017;23(3):538–539. doi:10.3201/eid2303.160833
9. Cameron DR, Jiang JH, Hassan KA, et al. Insights on virulence from the complete genome of Staphylococcus capitis. Front Microbiol. 2015;6:980. doi:10.3389/fmicb.2015.00980
10. Institute CaLS. Performance Standards for Antimicrobial Susceptibility Testing. 2018:M100
11. Waterhouse A, Bertoni M, Bienert S, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–W303. doi:10.1093/nar/gky427
12. Sander P, Springer B, Prammananan T, et al. Fitness cost of chromosomal drug resistance-conferring mutations. Antimicrob Agents Chemother. 2002;46(5):1204–1211. doi:10.1128/AAC.46.5.1204-1211.2002
13. Guo B, Abdelraouf K, Ledesma KR, Nikolaou M, Tam VH. Predicting bacterial fitness cost associated with drug resistance. J Antimicrob Chemother. 2012;67(4):928–932. doi:10.1093/jac/dkr560
14. van der Maten E, de Jonge MI, de Groot R, van der Flier M, Langereis JD. A versatile assay to determine bacterial and host factors contributing to opsonophagocytotic killing in hirudin-anticoagulated whole blood. Sci Rep. 2017;7(1):42137. doi:10.1038/srep42137
15. Tsai CJ-Y, Loh JMS, Proft T. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence. 2016;7(3):214–229. doi:10.1080/21505594.2015.1135289
16. Sun F, Li C, Jeong D, Sohn C, He C, Bae T. In the Staphylococcus aureus two-component system sae, the response regulator SaeR binds to a direct repeat sequence and DNA binding requires phosphorylation by the sensor kinase SaeS. J Bacteriol. 2010;192(8):2111–2127. doi:10.1128/JB.01524-09
17. Mehta AP, Hanes JW, Abdelwahed SH, Hilmey DG, Hanzelmann P, Begley TP. Catalysis of a new ribose carbon-insertion reaction by the molybdenum cofactor biosynthetic enzyme MoaA. Biochemistry. 2013;52(7):1134–1136. doi:10.1021/bi3016026
18. Yokoyama K, Leimkuhler S. The role of FeS clusters for molybdenum cofactor biosynthesis and molybdoenzymes in bacteria. Biochim Biophys Acta. 2015;1853(6):1335–1349. doi:10.1016/j.bbamcr.2014.09.021
19. Wi YM, Greenwood-Quaintance KE, Brinkman CL, Lee JYH, Howden BP, Patel R. Rifampicin resistance in Staphylococcus epidermidis: molecular characterisation and fitness cost of rpoB mutations. Int J Antimicrob Agents. 2018;51(5):670–677. doi:10.1016/j.ijantimicag.2017.12.019
20. Wichelhaus TA, Schafer V, Brade V, Boddinghaus B. Molecular characterization of rpoB mutations conferring cross-resistance to rifamycins on methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1999;43(11):2813–2816. doi:10.1128/AAC.43.11.2813
21. Bassenden AV, Rodionov D, Shi K, Berghuis AM. Structural analysis of the tobramycin and gentamicin clinical resistome reveals limitations for next-generation aminoglycoside design. ACS Chem Biol. 2016;11(5):1339–1346. doi:10.1021/acschembio.5b01070
22. Becker K, Heilmann C, Peters G. Coagulase-negative staphylococci. Clin Microbiol Rev. 2014;27(4):870–926.
23. Dong Y, Speer CP, Glaser K. Beyond sepsis: staphylococcus epidermidis is an underestimated but significant contributor to neonatal morbidity. Virulence. 2018;9(1):621–633. doi:10.1080/21505594.2017.1419117
24. Al Hennawi HET, Mahdi EM, Memish ZA. Native valve Staphylococcus capitis infective endocarditis: a mini review. Infection. 2019.
25. Laurent F, Butin M. Staphylococcus capitis and NRCS-A clone: the story of an unrecognized pathogen in neonatal intensive care units. Clin Microbiol Infect. 2019;25(9):1081–1085. doi:10.1016/j.cmi.2019.03.009
26. Tevell S, Hellmark B, Nilsdotter-Augustinsson A, Soderquist B. Staphylococcus capitis isolated from prosthetic joint infections. Eur J Clin Microbiol Infect Dis. 2017;36(1):115–122. doi:10.1007/s10096-016-2777-7
27. Carter GP, Ussher JE, Da Silva AG, et al. Genomic analysis of multiresistant staphylococcus capitis associated with neonatal sepsis. Antimicrob Agents Chemother. 2018;62(11):11. doi:10.1128/AAC.00898-18
28. Butin M, Martins-Simoes P, Pichon B, et al. Emergence and dissemination of a linezolid-resistant Staphylococcus capitis clone in Europe. J Antimicrob Chemother. 2017;72(4):1014–1020. doi:10.1093/jac/dkw516
29. O’Neill AJ, Huovinen T, Fishwick CW, Chopra I. Molecular genetic and structural modeling studies of Staphylococcus aureus RNA polymerase and the fitness of rifampin resistance genotypes in relation to clinical prevalence. Antimicrob Agents Chemother. 2006;50(1):298–309. doi:10.1128/AAC.50.1.298-309.2006
30. Munck C, Gumpert HK, Wallin AI, Wang HH, Sommer MO. Prediction of resistance development against drug combinations by collateral responses to component drugs. Sci Transl Med. 2014;6(262):262ra156. doi:10.1126/scitranslmed.3009940
31. Jacoby GA, Blaser MJ, Santanam P, et al. Appearance of amikacin and tobramycin resistance due to 4ʹ-aminoglycoside nucleotidyltransferase [ANT(4ʹ)-II] in gram-negative pathogens. Antimicrob Agents Chemother. 1990;34(12):2381–2386. doi:10.1128/AAC.34.12.2381
32. Miller GH, Sabatelli FJ, Hare RS, et al. The most frequent aminoglycoside resistance mechanisms–changes with time and geographic area: a reflection of aminoglycoside usage patterns? Aminoglycoside Resistance Study Groups. Clin Infect Dis. 1997;24(Suppl 1):S46–62. doi:10.1093/clinids/24.Supplement_1.S46
33. Nichol D, Rutter J, Bryant C, et al. Antibiotic collateral sensitivity is contingent on the repeatability of evolution. Nat Commun. 2019;10(1):334. doi:10.1038/s41467-018-08098-6
34. Imamovic L, Sommer MO. Use of collateral sensitivity networks to design drug cycling protocols that avoid resistance development. Sci Transl Med. 2013;5(204):204ra132. doi:10.1126/scitranslmed.3006609
35. Shapiro RS. mSphere of influence: evolutionary strategies to sensitize drug-resistant pathogens. mSphere. 2019;4:3. doi:10.1128/mSphere.00313-19
36. Kim S, Lieberman TD, Kishony R. Alternating antibiotic treatments constrain evolutionary paths to multidrug resistance. Proc Natl Acad Sci U S A. 2014;111(40):14494–14499. doi:10.1073/pnas.1409800111
37. Liu Q, Yeo WS, Bae T. The SaeRS Two-Component System of Staphylococcus aureus. Genes (Basel). 2016;7(10):10. doi:10.3390/genes7100081
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.