Back to Journals » International Journal of Nanomedicine » Volume 11
Effect of inserted spacer in hepatic cell-penetrating multifunctional peptide component on the DNA intracellular delivery of quaternary complexes based on modular design
Authors Zhang L, Li Z, Sun F, Xu YH , Du Z
Received 20 June 2016
Accepted for publication 20 September 2016
Published 24 November 2016 Volume 2016:11 Pages 6283—6295
DOI https://doi.org/10.2147/IJN.S115381
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Lei Yang
Luchen Zhang,1 Zhenbo Li,1 Fangli Sun,2 Yuhong Xu,1 Zixiu Du1
1School of Pharmacy, Shanghai Jiao Tong University, Shanghai, 2School of Engineering, Zhejiang Agriculture and Forestry University, Lin’an, Zhejiang Province, People’s Republic of China
Abstract: A safe and efficient quaternary gene delivery system (named Q-complexes) was constructed based on self-assembly of molecules through noncovalent bonds. This system was formulated through the cooperation and competing interactions of cationic liposomes, multifunctional peptides, and DNA, followed by coating hyaluronic acid on the surface of the ternary complexes. The multifunctional peptide was composed of two functional domains: penetrating hepatic tumor-targeted cell moiety (KRPTMRFRYTWNPMK) and a wrapping gene sequence (polyarginine 16). The effect of spacer insertion between the two domains of multifunctional peptide on the intracellular transfection of Q-complexes was further studied. Experimental results showed that the formulations assembled with various peptides in the spacer elements possessed different intercellular pathways and transfection efficiencies. The Q-complexes containing peptide in the absence of spacer element (Pa) showed the highest gene expression among all samples. The Q-complexes containing peptides with a noncleavable spacer GA (Pc) had no ability of intracellular nucleic acid delivery, whereas those with a cleavable spacer RVRR (Pd) showed moderate transfection activity. These results demonstrated that the different spacers inserted in the multifunctional peptide played an important role in in vitro DNA transfection efficiency. Atomic force microscopy images showed that the morphologies of ternary complexes (LPcD) and Q-complexes (HLcPD) were crystal lamellas, whereas those of other nanocomplexes were spheres. Circular dichroism showed the changed configuration of peptide with spacer GA in nanocomplexes compared with that of its free state, whereas the Pa configuration without spacer in nanocomplexes was consistent with that of its free state. The present study contributed to the structural understanding of Q-complexes, and further effective modification is in progress.
Keywords: Q-complexes, spacer elements, multifunctional peptides, lipid component, intracellular transfection
Introduction
Gene delivery can increase or decrease the expression of any protein in a cell theoretically,1 and thus drugs based on gene have been regarded a promising treatment for many serious diseases, such as cancer and genetic disorders.2,3 Effective gene delivery to the target sites remains a challenge because of the difficulty of transporting these negatively charged macromolecules to overcome various physical barriers without degradation.1,4 A safe and efficient delivery system is thus needed for a successful gene therapy.1,4,5 Since 1960s, nonviral vectors based on cationic polymers6–9 and lipids10 have been preferred as the carriers to wrap and transport genes.11 Numerous efforts have been made to optimize the designing strategies of nonviral vectors to overcome the barriers during delivery process.5 The conventional method of designing targeted drug carriers is via chemical conjugation of functional moieties;12 the conjugation of various ligands requires chemical reactions and may destroy the conformation of biomolecules, thereby resulting in inactivation and cytotoxicity. The self-assembly of molecules via noncovalent bonds to form an organized structure is most helpful in drug delivery system, maintains the nature of each component, and enhances the delivery activity of carriers through the synergies of all functional moieties.
On the basis of self-assembly of molecules via noncovalent bonds, we rationally constructed a highly effective and biologically responsive delivery system (Q-complexes) using nontoxic lipids (1,2-dioleoyl-3-trimethylammonium-propane [DOTAP] and 1,2-dioleoylsn-glycero-3-phosphatidyl-ethanolamine [DOPE]), peptide, and hyaluronic acid (HA). Briefly, the formulation is first composed of a cationic liposome, multifunctional peptide, and DNA at optimized ratios, which can finally self-assemble into ternary complexes (LPD). The ternary complex was associated with negatively charged HA and formulated quaternary complexes (HLPD, also named as Q-complexes). Specifically, we applied a hepatic tumor-specific cell-penetrating peptide (KRPTMRFRYTWNPMK)13 to polyarginine 16 (R16) in the absence or presence of spacer to create a multifunctional peptide, which could specifically permeate hepatic tumor cells and tightly condense nucleic acids. The HA coated at the surface of ternary complexes is in charge of shielding the negative charge of the nanoparticle surface, as well as the oriented binding to the tumor tissues and cells because HA actively targets receptors (such as CD44) distributed on the surfaces of the tumor cells.14,15 From the similar reported formulation,16 we could conclude that some of KRPTMRFRYTWNPMK in the multifunctional peptide protruded at the surface of LPD, which possibly induces the nanoparticles to penetrate into the tumor cells, and the lipid component destroys the endosomal membranes through membrane fusion function of DOPE.17
In the present study, we found that the transfection efficiency of HLPD is closely related to the inserted spacer of the multifunctional peptides, between KRPTMRFRYTWNPMK and R16. Thus, we investigated the effect of spacer element on the in vitro DNA transfection of HLPD and mainly explored the characteristics and internalization mechanism of HLPD containing the peptides with different spacers between KRPTMRFRYTWNPMK and R16. The cell uptake and gene expression of Q-complexes after incubation with different inhibitors were analyzed to validate the successful intracellular gene delivery pathway. In this text, H represents HA, L represents cationic liposome that was composed of DOTAP/DOPE at the weight ratio of 1:1, P represents peptide, and D represents DNA. For example, HLPaD refers to HA, cationic liposome, Pa, and DNA at the weight ratio of 14:1:4:1.
Materials and methods
Materials
DOTAP and DOPE were purchased from Sigma-Aldrich Co. (St Louis, MO, USA). The peptide sequence R16KMPNWTYRFRMTPRK (Pa), R16 (Pb), and peptide with the spacers GA and RVRR based on Pa (named as Pc and Pd) were purchased from China Peptides (Shanghai, People’s Republic of China) and dissolved in 10 mg/mL of nuclease-free water (Thermo Fisher Scientific, Waltham, MA, USA). The plasmid pCI-Luc comprising the luciferase gene of pGL3 was purchased from Invitrogen. The plasmids were amplified in DH5α strain of Escherichia coli and prepared using a GoldHi EndoFree Plasmid Maxi Kit (CWBiotech, Beijing, People’s Republic of China). HA (Mw =34 kDa) was purchased from Shandong Freda Biotechnology Co., Ltd. (Shandong, People’s Republic of China). In addition, fluorescein isothiocyanate-labeled HA (FITC-HA) was synthesized as reported.18
Preparation of cationic liposome
Liposome preparation was described previously.19 Briefly, the required amounts of DOTAP and DOPE were first dissolved in chloroform at the weight ratio of 1:1, and a thin lipid film was produced via slowly evaporating the chloroform overnight with a rotary evaporator under vacuum conditions. The lipid film was subsequently hydrated with double-distilled water at room temperature to produce a crude suspension of 1 mg/mL. The crude suspension was sonicated for 10 min to obtain the final product.
Formation of LPD and HLPD
Ternary complexes (LPD) were prepared by mixing the cationic liposome with different peptides and DNA solution at the weight ratio of 1:4:1 (ie, charge ratio of 0.3:4.4:1 for Pa, Pc, and Pd and charge ratio of 0.3:8.4:1 for Pb). These complexes were incubated for 30 min to formulate LPD at room temperature. The thoroughly mixed suspensions were mixed with HA solution at a precise ratio of 14:1 HA/DNA (ie, 5.9:1 charge ratio) and maintained for 15 min to produce Q-complexes (HLPD). For physicochemical studies of HLPD, the complexes were prepared and diluted with double-distilled water. For in vitro transfection, Opti-MEM was used as diluent.
Hydrodynamic size and zeta potential measurements
The hydrodynamic size and zeta potential of LPD and HLPD complexes were measured via dynamic light scattering using a Zetasizer Nano ZS (Malvern Instruments, Malvern, UK) at 25°C±0.1°C. The amount of DNA used in each formulation was 2 μg. The values for each formulation are the mean values of three measurements.
Gel electrophoresis assay
Approximately 1 μg of DNA and 10 μL of complex suspensions were used in the gel electrophoresis assay. All samples, except for 1 kb DNA ladder (Foregene, Chengdu, People’s Republic of China), were added to 2 μL of 6× loading buffer (Thermo Fisher Scientific) before loading the samples onto a 1% w/v agarose gel in Tris-acetate-EDTA buffer, which contained 3 μL of GelRed. The samples were electrophoresed at 90 V for 30 min and visualized under ultraviolet (UV) light.
Atomic force microscope
A drop of LPD and HLPD nanocomplexes was applied onto the mica. After a few seconds, the drop was dried through blotting with the filter paper. The total component concentration of each sample was 0.3 mg/mL. The morphology of LPD and HLPD was examined using an atomic force microscope (Nanoscope IIIa; Digital Instruments/Veeco, Inc., Santa Barbara, CA, USA).
Circular dichroism
Circular dichroism measurements were performed with aqueous preparations of LPD and HLPD complexes, as well as peptide and HA components at the same concentrations similar to those for the LPD and HLPD complexes. The individual components were used as controls to determine their native structures. All spectra were obtained in a JASCO J-815 spectropolarimeter using a 0.1 cm rectangular quartz cuvette cell, at a scan rate of 50 nm/min, data pitch of 1 nm, response time of 1 s, and bandwidth of 1 nm.
Cell culture
SMMC-7721 (human hepatoma) cells were obtained from Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Shanghai, People’s Republic of China). Cell culture media and reagents were purchased from Thermo Fisher Scientific. SMMC-7721 cells were cultured in RPMI-1640 medium. The medium used for cell culture was supplemented with 10% v/v fetal bovine serum and 1% v/v penicillin/streptomycin solution. All cell lines were incubated at 37°C in a humidified atmosphere with 5% CO2.
Flow cytometry and confocal laser scanning microscopy
DiI-dyed lipid component was used to monitor the trafficking of nanocomplexes. The cell uptake of the complexes was quantitatively measured via flow cytometry (FACS Calibur; BD, Worcestershire, NJ, USA). Cells were seeded in 24-well plates at the density of 1×105 cells per well and incubated at 37°C in a humidified atmosphere with 5% CO2 for 24 h before transfection. The culture medium was subsequently removed, and the cells were rinsed with 500 μL of 1× phosphate buffered saline (PBS). Afterward, the complexes containing 2 μg of pGL3 plasmid in 500 μL of Opti-MEM were added to each well. The cells were incubated at 37°C with 5% CO2 for 4 h, after which the complex suspensions were removed. The cells were collected and prepared for flow cytometry assay. For confocal microscopy, fluorescent-labeled Q-complex formulations were prepared with DOTAP and DOPE at 1:1 weight ratio containing DiI at 1.0% weight ratio of total lipids. In this experiment, the cells were incubated for 4 h. The cells were mixed with 4% paraformaldehyde, permeabilized with 0.3% triton in PBS for 10 min, and incubated for 5 min at room temperature with DAPI (0.2 mg/mL; Sigma-Aldrich). The cells were washed with PBS and sealed in mounting media (Invitrogen). Subsequently, visualization was carried out on a Leica TCS SP8 laser scanning microscope system (Leica Microsystems, Wetzlar, Germany).
Expression of green fluorescent protein (GFP) reporter gene
SMMC-7721 cells were seeded in 24-well plates at the density of 1×105 cells per well and incubated at 37°C in a humidified atmosphere with 5% CO2 for 24 h before transfection. The cells were first rinsed with 500 μL of 1× PBS, and the complexes containing 1 μg of GFP plasmid in 500 μL of Opti-MEM were added to each well. The cells were incubated at 37°C with 5% CO2 for 4 h. The suspensions were subsequently replaced with fresh medium, and the cells were incubated for additional 20 h. The cells were observed under a fluorescence microscope.
Intracellular internalization mechanism assay of HLPD
Cells were seeded in 96-well plates at the density of 2×104 cells per well and incubated at 37°C in a humidified atmosphere with 5% CO2 for 24 h prior to transfection. The culture medium was removed, and the cells were rinsed with 200 μL of 1× PBS. The complexes containing 250 ng of pGL3 in 200 μL of Opti-MEM were added to each well. The cells were incubated at 37°C with 5% CO2 for 4 h. Afterward, the complex suspensions were replaced with fresh complete culture medium, and the cells were incubated under the same condition for a further 20 h. Cells were then lysed with Reporter Lysis Buffer (Promega, Southampton, UK). A Luciferase Assay System (Promega) was used to measure luciferase activity with an illuminometer (Berthold Technologies, Bad Wildbad, Germany). The amount of protein in each lysate was determined with a Micro BCA Protein Assay Kit (Thermo Scientific) using a microplate reader (Bio-Rad Laboratories Inc., Hercules, CA, USA). Luciferase activity was expressed as relative light units per milligram of protein (RLU/mg protein). Moreover, four endocytic inhibitors, namely, dynamin inhibitor (DI: dynasore; Sigma-Aldrich), CD44-antibody (Abcam, Cambridge, MA, USA), chlorpromazine (CPZ, Aladdin, People’s Republic of China), and 5-(N-ethyl-N-isopropyl) amiloride (EIPA) were used to impede the dynamin-dependent, CD44-mediated, and macropinocytosis pathways. Cells were pretreated with endocytic inhibitors before transfection. For dynamin inhibition, the cells were first rinsed with 1× PBS, and the cells were subsequently incubated for 1 h at 37°C with 80 μM dynasore, 0.2% DMSO only, or the culture medium alone. Similarly, cells were incubated with anti-CD44 antibody (rabbit monoclonal [EPR1013Y] to CD44; 1/250), CPZ (20 μM, 1 h incubation), and EIPA (100 μM, 30 min incubation) with 0.1% DMSO at 37°C. After inhibitor pretreatment, the complexes were added to transfect cells. A luciferase activity assay was then conducted with the same procedures as mentioned earlier. For flow cytometric analysis, cells were incubated for 4 h in the presence or absence of endocytosis inhibitors of dynasore or CD44 antibody. Cell uptake was measured via flow cytometry (FACS Calibur).
Statistical analysis
All experiments were performed in triplicate, and the data are represented as mean ± SD. Student’s t-tests were used for statistical analysis among groups. A value of *P<0.05 was considered significant. All the analyses were conducted using GraphPad Prism 5 Instat software (GraphPad, San Diego, CA, USA).
Results
Physicochemical characteristics of ternary and Q-complexes
The amino acid sequences of multifunctional peptides are shown in Table 1. These sequences were used as the peptide component for investigation of the effect of spacer in peptide on the characteristics and delivery efficiency of Q-complexes. The hydrodynamic size and zeta potential of the LPD and HLPD complexes studied are illustrated in Figure 1. The complexes except for LPcD were basically ~100 nm in diameter with appropriate positive charge for LPD in the range of 20–50 mV. For HLPD, the values were reversed to negative charge ranging from −30 mV to −40 mV of zeta potential. The zeta potential of HLPcD was −19 mV at the weight ratio of 1:4:1. Therefore, we further investigated the formulation of Q-complexes based on Pc. The small self-assembled structure of ternary complexes was obtained at 8:1 weight ratio of Pc:D, and the size and zeta potential of LPD and HLPD were found to be similar (Figure S1) at 4:1 weight ratio of Pc:D. The gel electrophoresis results indicated that all complexes, except the samples based on Pc, were able to condense nucleic acids effectively at their optimized formulations (Figures 2 and S2), whereas LPcD and HLPcD showed poor DNA condensation ability. The atomic force microscope images (Figure 3) showed that LPaD and HLPaD presented uniform spheres. The ternary and Q-complexes containing Pb and Pd also formed spherical nanoparticles with various sizes. However, LPcD and HLPcD formed crystal lamellas, which were considerably different from those of other samples.
Circular dichroism spectra
To investigate the alteration in the conformation of peptides in the complexes, the circular dichroism of Q-complexes (HLPaD, HLPcD, and HLPdD) was examined in the range of 190–300 nm (Figure 4). Responsive ternary complexes (LPaD, LPcD, and LPdD), peptides (Pa, Pc, and Pd), and free HA were also examined. The circular dichroism signal of free liposome was not measured because of the absence of chromophores absorbing in the UV region.17 As shown in Figure 4A, the magnitude of negative circular dichroism band had a λmax at ~208 nm in individual Pa and LPaD. The λmax of free HA was located at 210 nm, and the peak of HLPaD was the overlap of Pa and HA component peaks. The abovementioned results demonstrated that Pa showed an unchanged α-helix configuration in a free state, which was the same as in LPaD and HLPaD. Furthermore, the λmax at ~208 nm appeared in free Pc, but moved to 210 nm in LPcD and HLPcD, and also appeared in a positive peak at ~195 nm in LPcD and HLPcD. The result indicated that the secondary structure of Pc had changed from α-helix toward β-sheet (Figure 4B). The negative λmax of peptide in LPdD and free Pd at 200 nm indicated the random coil, and the λmax of peptide in HLPdD was at 218 nm (Figure 4C), which showed that the secondary structure of Pd in HLPdD was the transition state from random coil into β-sheet.
Cell uptake
The Q-complexes containing Pa, Pb, and Pd, respectively, were effectively internalized after 4 h of transfection (Figure 5). The equivalent mean fluorescence intensity of the cells after incubation with Q-complexes and the corresponding ternary complexes could be observed from the cell uptake transfected to SMMC-7721 cells. Such observation indicated that the internalization efficiency of nucleic acids did not drop after coating the surface of ternary complexes with HA. However, the Q-complexes containing Pc were not easily endocytosed by the tumor cells.
EGFP expression
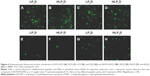
Fluorescence microscope was used to observe the GFP expression in varying intensities after incubation with SMMC-7721 cells for 24 h. The HLPaD sample showed the highest transfection activity (Figure 6) in all the samples containing Pa–Pd. Specifically, the GFP expression efficiency of complexes was in the order of samples containing the following peptides: Pa > Pb > Pd > Pc. The GFP expression results indirectly reflected the DNA delivery trend of Q-complexes with different multifunctional peptides.
Luciferase transfection and internalization pathways of ternary and Q-complexes
SMMC-7721 cells were transfected with LPD and HLPD complexes for the investigation of both transfection efficiency and internalization pathways of Q-complexes. Figure 7 illustrates that the intercellular activity of the ternary and Q-complexes containing Pa showed the highest transfection efficiency among all the samples in the absence of inhibitors, and the complexes containing Pb showed lower transfection activity compared with that of complexes containing Pa. On the contrary, the complexes containing Pc barely delivered DNA into the cells. The samples containing Pd showed higher DNA transfection efficiency than those complexes containing Pc.
The unchangeable transfection efficiencies of all complexes could be observed after incubation with CD44 antibody (Figure 7A), which indicated the minimal contribution of CD44-mediated internalization to the intercellular transfection activity of the complexes. In addition, a significant drop in the luciferase expression of the samples containing Pa was observed after pretreatment with dynasore (Figure 7B). This result showed that the internalization pathway of these nanocomplexes was mainly dynamin-dependent, thereby reflecting a strong penetrating ability of the peptide component in the nanoparticles.13 The internalization mechanism of most LPD and HLPD was also partly both clathrin-20 (Figure 7C) and macropinocytosis-dependent21 (Figure 7D) pathways.
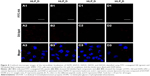
We further investigated the intracellular paths of Q-complexes observed through confocal microscopy (Figure 8). FITC-HA and DiI-labeled lipid component (red) were used to monitor the trafficking of HLPD inside SMMC-7721 cells. Nuclei were stained with DAPI (blue). After incubation for 4 h, the HLPaD and HLPbD inside the SMMC-7721 cells were observed through DiI-labeled lipid component, but the dots of FITC-HA were not found. This result implied that HA component had separated from the remaining components of Q-complexes before termination of incubation. Similarly, it also indicated that the FITC-HA of Q-complexes could only be efficiently endocytosed through CD44-mediated pathway (Figure 9). We also found that anti-CD44 antibody impeded the fluorescence intensity of FITC-HA, and dynasore could not prevent the cell uptake of FITC-HA. Therefore, HLPcD could not be endocytosed by the SMMC-7721 cells.
Discussion
Multifunctional peptide consists of both nearly neutral (KRPTMRFRYTWNPMK) and cationic peptide (R16) sequences. Thus, the effects of incorporating peptide between these two peptide sequences are important for rationally designing a delivery system. A large gap of transfection efficiency between HLPaD and HLPcD inspired us to investigate the role of the incorporating spacer in the peptide component of Q-complexes. We selected two representative peptide spacers (GA and RVRR) as different spacer elements based on the original peptide of Pa (Table 1) and verified whether these changes would affect the formulation and transfection efficiency of Q-complexes. In addition, we inhibited the intercellular uptake pathways, mainly including the CD44 mediating and dynamin-dependent pathways that HA and the tumor cell-penetrating peptide (KMPNWTYRFRMTPRK) specifically depended on,13 respectively. Q-complexes were hypothesized to achieve intracellular delivery through multifunctional peptide completely wrapping nucleic acids and strongly penetrating specific tumor cell lines. Consequently, we employed DOPE lipid component to disrupt the endosomal membrane, in addition to the negatively charged HA coating at the surface of ternary complexes to shield the positive surface charges and highly effective targeting tumor. The experimental results showed that the nanocomplexes containing different peptide components possessed similar sizes (Figure 1), and the zeta potential of LPcD was negative at approximately −19 mV; other LPD formulations had positive surface charge. The electrophoretic mobility results (Figure 2) indicated that LPcD and HLPcD could not tightly wrap DNA. In contrast to other spherical nanoparticles, the morphology and structure of LPcD and HLPcD obviously changed to crystal lamellas, which perhaps implied the reason why the samples containing Pc had bare DNA transfection efficiency. The circular dichroism result (Figure 4) further demonstrated that folding or conformational changes in Pc integrated into LPcD and HLPcD caused the minimal cell uptake and almost failure in transport DNA ability of the delivery system. Taking the results of cell uptake (Figure 5) and transfection efficiencies (Figures 6 and 7) into account, we thought that the reason why complexes with Pc showed low efficiency might be that undesirable formation could not be changed immediately in the transport process as GA is a noncleavable spacer. Additionally, when RVRR instead of GA was used as a spacer in Pd, the morphologies of nanocomplexes were still spherical, and the conformational change of Pd occurred in HLPdD but not in LPdD. Therefore, different inserted spacer elements of peptides could result in different morphologies and transfection efficiencies of LPD and HLPD formations. The enhanced cellular uptake and transfection efficiency of LPdD and HLPdD demonstrated that the negative effect of RVRR on the intracellular delivery efficiency was lower than that of GA. Thus, the crystal morphologies of complexes and some conformational changes in multifunctional peptides might shield their penetrating function or weaken their penetrating ability. However, when the multifunctional peptide was formed without spacer, the strong cell-penetrating function of KRPTMRFRYTWNPMK and tightly wrapping nucleic acid capability of R16 could be fully manifested in LPaD and HLPaD. Moreover, the LPaD and HLPaD formulations showed unified spherical morphology, and the Pa in the ternary and Q-complexes maintained unchangeable configuration. The function of HA was mainly in shielding the positive charge and pushing the nanoparticles to bind to the tumor cells. LPaD and HLPaD transport DNA mainly through dynamin-dependent and CD44-mediated independent pathways (Figure 6), which is completely different from the traditional drug delivery system utilizing HA for both targeting CD44 receptor and entering the cell through CD44-mediated pathway.15,22,23
Conclusion
We rationally designed a stable delivery platform for in vivo nucleic acid delivery based on modular design, which integrates a specific tumor lineage-homing cell-penetrating peptide and gene-condensing peptide into a multifunctional peptide that can both envelope nucleic acids and penetrate the tumor cell membrane. This study has proven that spacers in the peptide could decrease and prevent the intracellular transfection efficiency through reversing the complexes morphology from sphere to crystal lamellae and changing the peptide configuration. These findings will be important in understanding the formulation of Q-complexes and beneficial in designing an improved gene delivery system in the future.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No 30901881 and 31470587) and Engineering and Medical Cooperation Projects of Shanghai Jiao Tong University (No YG2013MS42 and YG2014QN04).
Disclosure
The authors report no conflicts of interest in this work.
References
Nguyen J, Szoka FC. Nucleic acid delivery: the missing pieces of the puzzle? Acc Chem Res. 2012;45(7):1153–1162. | ||
Ibraheem D, Elaissari A, Fessi H. Gene therapy and DNA delivery systems. Int J Pharm. 2014;459(1–2):70–83. | ||
Yang ZR, Wang HF, Zhao J, et al. Recent developments in the use of adenoviruses and immunotoxins in cancer gene therapy. Cancer Gene Ther. 2007;14(7):599–615. | ||
Guo X, Huang L. Recent advances in nonviral vectors for gene delivery. Acc Chem Res. 2012;45(7):971–979. | ||
Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Non-viral vectors for gene-based therapy. Nat Rev Genet. 2014;15(8):541–555. | ||
Morris VB, Labhasetwar V. Arginine-rich polyplexes for gene delivery to neuronal cells. Biomaterials. 2015;60:151–160. | ||
Lu S, Morris VB, Labhasetwar V. Codelivery of DNA and siRNA via arginine-rich PEI-based polyplexes. Mol Pharm. 2015;12(2):621–629. | ||
Khargharia S, Kizzire K, Ericson MD, Baumhover NJ, Rice KG. PEG length and chemical linkage controls polyacridine peptide DNA polyplex pharmacokinetics, biodistribution, metabolic stability and in vivo gene expression. J Control Release. 2013;170(3):325–333. | ||
Olins DE, Olins AL, Von Hippel PH. Model nucleoprotein complexes: studies on the interaction of cationic homopolypeptides with DNA. J Mol Biol. 1967;24(2):157–176. | ||
Marimani MD, Ely A, Buff MC, et al. Inhibition of replication of hepatitis B virus in transgenic mice following administration of hepatotropic lipoplexes containing guanidinopropyl-modified siRNAs. J Control Release. 2015;209:198–206. | ||
Sitharaman B, Zakharian TY, Saraf A, et al. Water-soluble fullerene (C60) derivatives as nonviral gene-delivery vectors. Mol Pharm. 2008;5(4):567–578. | ||
Kozielski KL, Rui Y, Green JJ. Non-viral nucleic acid containing nanoparticles as cancer therapeutics. Expert Opin Drug Deliv. 2016;13(10):1475–1487. | ||
Kondo E, Saito K, Tashiro Y, et al. Tumour lineage-homing cell-penetrating peptides as anticancer molecular delivery systems. Nat Commun. 2012;3:951. | ||
Stern R. Hyaluronan metabolism: a major paradox in cancer biology. Pathol Biol. 2005;53(7):372–382. | ||
Yamada Y, Hashida M, Harashima H. Hyaluronic acid controls the uptake pathway and intracellular trafficking of an octaarginine-modified gene vector in CD44 positive- and CD44 negative-cells. Biomaterials. 2015;52:189–198. | ||
Mustapa MF, Bell PC, Hurley CA, et al. Biophysical characterization of an integrin-targeted lipopolyplex gene delivery vector. Biochemistry. 2007;46(45):12930–12944. | ||
Munye MM, Ravi J, Tagalakis AD, McCarthy D, Ryadnov MG, Hart SL. Role of liposome and peptide in the synergistic enhancement of transfection with a lipopolyplex vector. Sci Rep. 2015;5:9292. | ||
Hua Q, Knudson CB, Knudson W. Internalization of hyaluronan by chondrocytes occurs via receptor-mediated endocytosis. J Cell Sci. 1993;106(pt 1):365–375. | ||
Du Z, Munye MM, Tagalakis AD, Manunta MD, Hart SL. The role of the helper lipid on the DNA transfection efficiency of lipopolyplex formulations. Sci Rep. 2014;4:7107. | ||
Marina-Garcia N, Franchi L, Kim YG, et al. Clathrin- and dynamin-dependent endocytic pathway regulates muramyl dipeptide internalization and NOD2 activation. J Immunol. 2009;182(7):4321–4327. | ||
Nakase I, Tadokoro A, Kawabata N, et al. Interaction of arginine-rich peptides with membrane-associated proteoglycans is crucial for induction of actin organization and macropinocytosis. Biochemistry. 2007;46(2):492–501. | ||
Chono S, Li SD, Conwell CC, Huang L. An efficient and low immunostimulatory nanoparticle formulation for systemic siRNA delivery to the tumor. J Control Release. 2008;131(1):64–69. | ||
He Y, Cheng G, Xie L, Nie Y, He B, Gu Z. Polyethyleneimine/DNA polyplexes with reduction-sensitive hyaluronic acid derivatives shielding for targeted gene delivery. Biomaterials. 2013;34(4):1235–1245. |
Supplementary materials
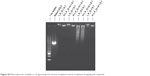  | Figure S2 Electrophoretic mobility in 1% agarose gel for ternary complexes and Q-complexes wrapping pGL3 plasmid. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.