Back to Journals » International Journal of Nanomedicine » Volume 15
Effect of Graphene Oxide and Silver Nanoparticles Hybrid Composite on P. aeruginosa Strains with Acquired Resistance Genes
Authors Lozovskis P, Jankauskaitė V , Guobienė A , Kareivienė V, Vitkauskienė A
Received 23 October 2019
Accepted for publication 28 May 2020
Published 17 July 2020 Volume 2020:15 Pages 5147—5163
DOI https://doi.org/10.2147/IJN.S235748
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Anderson Oliveira Lobo
Povilas Lozovskis,1 Virginija Jankauskaitė,2 Asta Guobienė,3 Violeta Kareivienė,1 Astra Vitkauskienė1
1Faculty of Medicine, Lithuanian University of Health Science, Kaunas, Lithuania; 2Department of Production Engineering, Kaunas University of Technology, Kaunas, Lithuania; 3Institute of Materials Science, Kaunas University of Technology, Kaunas, Lithuania
Correspondence: Povilas Lozovskis
Faculty of Medicine, Lithuanian University of Health Science, Mickevičiaus St. 9, Kaunas LT-44307, Lithuania
Tel +37060594939
Email [email protected]
Background: In the last decades, nosocomial infections caused by drug-resistant Pseudomonas aeruginosa became a common problem in healthcare facilities. Antibiotics are becoming less effective as new resistant strains appear. Therefore, the development of novel enhanced activity antibacterial agents becomes very significant. A combination of nanomaterials with different physical and chemical properties enables us to generate novel multi-functional derivatives. In this study, graphene oxide and polyvinylpyrrolidone-stabilized silver nanoparticles hybrid nanocomposite (GO-Ag HN) were synthesized. The relation between antibiotic resistance and GO-Ag HN potential toxicity to clinical P. aeruginosa strains, their antibiotic resistance, and molecular mechanisms were assessed.
Methods: Chemical state, particle size distribution, and morphology of synthesized GO-Ag NH were investigated using spectroscopy and microscopy techniques (UV-Vis, FTIR, XPS, TEM, SEM, AFM). Broad-spectrum antibiotic resistance of P. aeruginosa strains was determined using E-test. Antibiotic resistance genes were identified using polymerase chain reaction (PCR).
Results: In this study, the toxicity of the GO-Ag NH to the isolated clinical P. aeruginosa strains has been investigated. A high antibiotic resistance level (92%) was found among P. aeruginosa strains. The most prevalent antibiotic resistance gene among tested strains was the AMPC beta-lactamase gene (65.6%). UV-vis, FTIR, and XPS studies confirmed the formation of the silver nanoparticles on the GO nanosheets. The functionalization process occurred through the interaction between Ag nanoparticles, GO, and polyvinylpyrrolidone used for nanoparticle stabilization. SEM analysis revealed that GO nanosheets undergo partial fragmentation during hybrid nanocomposite preparation, which remarkably increases the number of sharp edges and their mediated cutting effect. TEM analysis showed that GO-Ag HN spherical Ag nanoparticles mainly 9– 12 nm in size were irregularly precipitated on the GO nanosheet surface. A higher density of Ag NPs was observed in the sheets’ wrinkles, corrugations, and sharp edges. This hybrid nanocomposite poses enhanced antibacterial activity against carbapenem-resistant P. aeruginosa strains through a possible synergy between toxicity mechanisms of GO nanosheets and Ag nanoparticles. With incubation time increasing up to 10 minutes, the survival of P. aeruginosa decreased significantly.
Conclusion: A graphene oxide and silver nanoparticles hybrid composite has been shown to be a promising material to control nosocomial infections caused by bacteria strains resistant to most antibiotics.
Keywords: graphene oxide nanosheets, silver nanoparticles, hybrid nanocomposite, antibacterial activity, Pseudomonas aeruginosa, carbapenem-resistant bacteria
Introduction
The situation in clinical microbiology has changed drastically over the last decades. The widespread use of broad-spectrum antibiotics has generated conditions for the appearance of multidrug-resistant bacteria that further cause clinical problems.1 Bacteria have developed many ways of antibiotic resistance, such as decreased cell permeability, enzyme inactivation, target protection and overproduction, and increased efflux due to over-expression.2 Nosocomial infections cause
d by previously uncommon bacteria became a major issue in hospitals worldwide. Pseudomonas aeruginosa is one of the most common opportunistic pathogens to cause nosocomial infections.3 The main feature that allowed P. aeruginosa strains to become a major nosocomial pathogen was its resistance. P. aeruginosa has already been shown to have intrinsic resistance mechanisms like beta-lactamase production, up-regulated efflux pumps, and reduced permeability of the outer membrane that reduces susceptibility to aminoglycosides, fluoroquinolones, and beta-lactams.4,5 Moreover, P. aeruginosa strains have acquired a broad spectrum of antibiotic resistance genes. The most notable of them belong to the class B beta-lactamases, like blaVIM, blaIMP, blaGIM, or blaSPM.6,7 These mechanisms allow P. aeruginosa not only to survive in hospitals but also to thrive and reproduce.
Antibiotics are becoming less effective as new resistant strains appear, limiting treatment options.8 The importance of preventing the infection is becoming clearer as the costs of treatment grow.9 Therefore, there is a great significance and importance to develop alternate antibiotic-independent drugs. In this respect, nanomaterials are promising due to their unique physical and chemical properties, such as large surface area relative to volume, that enables intimate interaction with microorganism’s membrane, biocompatibility, surface functionalization, and which helps in the development of more effective antibacterial agents.10–12 Moreover, the antibacterial nanomaterials can overcome the problem of resistance and diminish undesirable side effects of the treatment, to some extent owing to the use at a much lower dose.14
Broad-spectrum antibacterial properties against both Gram-positive and Gram-negative bacteria of metal-based nanoparticles (NPs) have been demonstrated.11,13 Long-term antibacterial and biofilm prevention provided by NPs is correlated with the structural and physical properties such as size, shape, surface charge, concentration, and colloidal state.15 The properties of NPs can be controlled by choosing the method of synthesis and varying reaction conditions such as the precursor concentration, molar ratio of the surfactant, etc.16 Silver nanoparticles (Ag NPs) can be prepared by physical methods, chemical synthesis, or using biological techniques.16,17 They are the most effective nano-agent against bacterial infections, allowing their use in a wide range of applications. Ag NPs seem to have high potential to solve the problem of multidrug-resistance, which is often observed in bacterial strains like Acinetobacter baumannii, Klebsiella pneumonia, and Pseudomonas aeruginosa.18–20 The toxicity of Ag NPs is a concentration and time dependent.20 Although Ag NPs are effective against more than 650 pathogens, the precise mechanism of their antimicrobial action is not fully understood yet.21 Nevertheless, Ag NP adhesion to microbial cells, penetration through the cell wall, generation of reactive oxygen forms and free radicals, and modulation of microbial signal transduction pathways, have been recognized as the most prominent modes of antimicrobial action that results in cell death.21,22 The combined bactericidal effect of the released Ag+ ions and Ag NPs may be possible.23 Unfortunately, Ag NPs lose their active surface area and show weaker antibacterial activity due to the spontaneous aggregation of nanoparticles. To overcome this problem, a carrier that disperses Ag NPs and enhances their antibacterial effects could be used. At this point, a graphene-based material, ie, graphene oxide (GO), seems to be a reliable nanomaterial due to easy processing, low cost of production, unique chemical and physical properties, and ability to kill or inhibit bacteria on its own.24–27 Recent studies show that the antibacterial efficiency of GO is caused by its sharp edge-mediated cutting effect, cell entrapment ability, and oxidative stress effect.28,29 However, some negative results of the GO antibacterial effect on P. aeruginosa have been found. They are related to the negative charge of the bacterial cell membrane or the periplasmic space that leads to a tendency to repulse the mechanical interactions between GO and Gram-negative bacteria.30,31 Meanwhile, the GO-based nanocomposites with metal nanoparticles have shown strong inhibition efficiency against nosocomial infection.32,33 It is reported that metal nanoparticles can interact with the GO sheets through electrostatic binding, physisorption, and charge–transfer interactions. Nanocomposites composed of GO and Ag NPs are mainly fabricated using various reductants, such as sodium borohydride, sodium citrate, hydrazine monohydrate, ascorbic acid, glucose, starch, hydroquinone, microorganisms, and plant extracts.27,34,35 GO surface modification and functionalization with Ag NPs enhance antimicrobial activity, and increase stability and dispersity.32,34,35 Additionally to the synergistic antibacterial effect, GO can prevent the aggregation of Ag NPs, and, in turn, the Ag NPs, immobilized on the GO nanosheet surface, prevent the aggregation of GO.32 Zhu et al36 reported that an increase of the GO-Ag NP-based nanocomposite antibacterial effect could also be attributed to the large amount of cations on the surface of the composite, which enhances the interaction with bacterial cell membranes.
Despite several achievements, the contribution of each physicochemical property on the antibacterial activity of graphene-based nanocomposites is still not clear. Therefore, considerable efforts are needed to understand the factors affecting the interactions and mechanisms involved in induced bacterial death. Besides, the antibacterial tests on the majority of graphene-based nanocomposites were applied mainly against E. coli and S. aureus. Therefore, it is very important to test other pathogenic species to demonstrate a broad bactericidal range of hybrid nanocomposites. In this study, the antibiotic resistance of isolated clinical P. aeruginosa strains was evaluated and toxicity of the graphene oxide and silver nanoparticles hybrid composite on these pathogenic bacteria has been investigated.
Materials and Methods
Materials
Highly concentrated graphene oxide (GO) aqueous dispersion with a concentration of 5 mg/mL (carbon – 79%, oxygen – 20%) and flake size of 0.5–5 μm was used as received (from Graphene Laboratories Inc.). Silver nitrate (AgNO3, ≥99.8% Sigma-Aldrich), polyvinylpyrrolidone with a molecular weight of Mw = 40,000 mol−1 (PVP40, Sigma-Aldrich) and other used chemicals were of analytical grade.
Preparation of GO-Ag Nanocomposite
Firstly, the Ag NP colloidal solution in PVP was prepared by the chemical reduction method. In this case, the appropriate amount (2 g) of PVP, used as reducing agent, stabilizer, and anticoagulant, was dissolved in ethanol (8 g). An AgNO3 solution was added to deionized water (0.2 g AgNO3 in 1 mL H2O) drop-wise with vigorous stirring at room temperature for 1 h (AgNO3:PVP=1:10). PVP-stabilized Ag NPs were formed and their colloidal solution of ca. 10 mg/mL concentration was obtained after precipitation of 24 h at room temperature. After this period, the solution color changed from light yellow to yellowish-brown.
The GO and PVP-stabilized Ag NPs hybrid nanocomposite (GO-Ag HN) was prepared by drop-wise addition of an Ag NP dispersion to the GO dispersion (5 mg/mL) at a ratio of GO:Ag NPs = 1.3:1. GO-Ag HN colloidal solution was sonified for 30 min to produce a homogeneous nanocomposite mixture with an Ag NP concentration of ca. 5 mg/mL. Figure 1 briefly shows the fabrication steps of the GO-Ag nanocomposite.
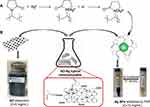 |
Figure 1 Illustration for the preparation of the GO-Ag hybrid nanocomposite: (A) synthesis of PVP-stabilized Ag NPs (interaction mechanism), (B) GO and Ag NPs solutions mixing scheme. |
Characterization of GO-Ag Nanocomposite
An optical spectrometer Avantes, composed of a deuterium halogen light source AvaLight DHc (Avantes, Apeldoorn, the Netherlands) and spectrometer Avaspec-2048 (Avantes, Apeldoorn, the Netherlands), was used to record UV-visible (UV-vis) light absorbance spectra. The analysis was performed in the wavelength range of 200–800 nm.
The Fourier transform infrared (FTIR) spectra were recorded using the Spectrum GX FTIR spectrometer (Perkin-Elmer, USA), equipped with a horizontal attenuated total reflection (HATR) accessory. The HATR FTIR spectra of samples were recorded at room temperature in the wavenumber range of 4000–600 cm−1 with a resolution of 1 cm−1. Collected spectra were processed with the Spectrum® v5.0.1 software from the Perkin-Elmer.
X-ray photoelectron spectroscopy (XPS) measurements were carried out to obtain information about the elemental chemical states and surface composition of GO and its nanohybrid samples on the upgraded Vacuum Generator (VG) ESCALAB MKII spectrometer fitted with a new XR4 twin anode. The non-monochromatized MgKα X-ray source was operated at hᶹ = 1253.6 eV with 300 W power (20 mA/15 kV). During spectral acquisition, pressure in the analysis chamber was lower than 5x10–7 Pa. The spectra were acquired using an electron analyzer pass energy of 20 eV for narrow scans and resolution of 0.05 eV, and with a pass energy of 100 eV for survey spectra. All spectra were recorded at a take-off angle of 90° and calibrated from the hydrocarbon contamination using the C 1s peak at 284.8 eV. The spectra calibration, processing, and fitting routines were performed using Avantage software (5.962) provided by Thermo VG Scientific (Waltham, MA, USA). Core level peaks of N 1s, Ag 3d, C 1s, S2p, and O 1s were analyzed using a nonlinear Shirley-type background. The calculation of the elemental composition Scofield’s based on relative sensitivity factors was performed.
Microscopy investigations were used for the GO nanohybrid morphology characterization. TEM images were acquired by Tecnai G2 F20 X-TWIN (FEI) equipped with a field emission electron gun. The TEM accelerating voltage was 200 kV. Elemental analysis was performed using an energy dispersive X-ray (EDX) spectrometer. Samples were prepared by diluting colloidal solutions of GO and its composite in ethanol and placing a drop of solution on a Lacey carbon grid and left overnight at ambient temperature.
Scanning electron microscopy (SEM) micrographs were acquired using field emission scanning electron microscope Quanta 200 FEG (FEI, Oregon, USA) and e-line plus multi-application nanoengineering workstation (Raith, Dortmund, Germany).
Surface topography imaging and detection of bacteria inhibition were carried out using atomic force microscopy (AFM). AFM experiments were carried out at room temperature using a NanoWizardIII microscope (JPK Instruments, Bruker Nano GmbH, Berlin, Germany), and data were analyzed using a SurfaceXplorer (Microtestmachines, Gomel, Belarus) and JPKSPM Data Processing software (Version spm-4.3.13, JPK Instruments, Bruker Nano GmbH, Berlin, Germany). The AFM images (scanning area 4×4 μm) were collected using an AppNano production V-shaped n-type silicon cantilever (0.01–0.025 ohm/cm, spring constant of 13–77 N/m, tip curvature radius of 10.0 nm and cone angle of 20°) operating in contact mode.
For SEM, AFM and XPS testing samples were prepared by spreading a thin layer of tested GO-based colloidal solutions on a silicon-covered chip and drying it at 35°C for 24 h.
Isolation of P. aeruginosa and the Antibiotic Susceptibility Testing
A retrospective P. aeruginosa collection of 64 carbapenem-resistant clinical strains was tested. Samples were collected during the 2011–2013 period and kept in a freezer at –80°C temperature. Antibiotic resistance was determined using the E-test (Liofilchelm, Italy) method, and interpreted by EUCAST (European Committee on Antimicrobial Susceptibility Testing) guidelines. For molecular testing of resistance genes DNA was extracted using QIAamp DNA Mini Kit (QIAGEN, USA) and PCR was run with specific primers as described previously.37 Gel electrophoresis was run in 2% agarose gel to visualize PCR results.
GO-Ag HN Antibacterial Assay
To determine the bacterial growth inhibition of GO-Ag hybrid nanocomposite to carbapenem- resistant bacteria we used previously developed methodology.38 P. aeruginosa strains were cultivated overnight on 5% blood agar (BD, USA) at a temperature of 37°C. Saline was used to make P. aeruginosa inoculate of 0.5 MF concentration. Nine parts of inoculate were mixed with one part of the GO-Ag hybrid nanocomposite solution. The mix was pulse vortexed and spread on 5% blood agar plate with a 1 µL loop instantly after vortexing, and every 1 min interval up to 10 min, and then after 120 min. Bacterial growth was evaluated after plates were incubated for 24 h at 37°C.
Statistical Analysis
For structural and morphological analysis, reported values refer to an average of not less than three tests. For bacteria susceptibility testing, statistical significance was determined using IBM SPSS statistics (IBM, USA) software. Statistical analysis was performed using the Chi-squared test. To determine results as significant p<0.05 was used.
Results
Characteristics of the GO-Ag Nanocomposite
UV-vis spectroscopy was used to monitor the formation of Ag NPs on GO sheets. Two characteristic bands were observed in the UV-vis spectrum of pristine GO (Figure 2). An absorption band at 227 nm is attributed to π–π* transitions of the aromatic C–C bonds, while the shoulder centered at 302 nm is derived from n–π* transitions of C=O bonds. Meanwhile, the UV-vis spectrum of Ag NPs contains a strong absorption peak centered at 425 nm due to the localized surface plasmon resonance (LSPR) of nanoparticles. It can be proposed that Ag NPs with a mean size of 25 nm are synthesized after the reduction of AgNO3 in the presence of PVP, whereas the UV peak position depends on the mean size of nanoparticles.39,40 The attachment of Ag NPs to the GO surface is confirmed primarily through the detection of a damped absorption band attributed to the LSPR of nanoparticles. Additionally, this absorption band is blue-shifted to 414 nm, indicating that GO nanosheets are functionalized with Ag NPs of smaller size. Moreover, the redshift of the GO absorption peak at 227–258 nm in the GO-Ag HN spectrum demonstrates that the oxygen-containing functional groups on the GO were mostly removed and partial restoration of π–conjugation within the graphene sheets occurred via reduction reaction and the formation of the nanohybrid structure.41,42
 |
Figure 2 UV-vis absorption spectra of GO (A), Ag NPs (B) and GO-Ag HN (C). |
FTIR measurements were carried out to investigate the interaction between GO nanosheets and Ag NPs. As can be seen in Figure 3, GO exhibits strong absorption peaks related to the stretching vibrations of OH (3396 cm−1), C=O in carboxyl and carbonyl moieties (1711 cm−1), skeletal vibration of aromatic C=C or intramolecular hydrogen bonds or residual H2O (1621 cm−1).43,44 The other peaks at 1387 cm−1, 1222 cm−1, and 1053 cm−1 correspond to C–O–H deformation, C–H stretching (epoxy groups), and C–O–C stretching (alkoxy groups) vibrations, respectively.45 Such an abundance of hydroxyl and oxygenous groups makes GO convenient for modification with plasmonic Ag NPs. FTIR spectrum of PVP-stabilized Ag NPs mainly displays absorption band characteristics for PVP.46,47 A strong peak at 1286 cm−1, typical of a –C–N bond, and a peak at 1635 cm–1 related to the carbonyl group C=O are observed in the Ag NP spectrum. Oxygen and nitrogen atoms arising from PVP units are involved in the formation of coordination bonds with silver atoms48 and GO.47 In the spectrum of GO-Ag HN, the carboxylic (C=C) and carbonyl (C=O) stretching vibrations of the GO carbon skeletal structure are redshifted. This can be attributed to the functionalization process that occurred through bonding between GO and PVP-stabilized Ag NPs. Besides, the changes of wavenumbers for the –OH stretching vibrations (from 3401 to 3447 cm−1) are usually related to hydrogen bonding formation.47
 |
Figure 3 FTIR spectra of GO, Ag NPs and GO-Ag HN. |
The chemical state of materials to be tested was investigated using XPS analysis. In Figure 4A the survey spectra clearly demonstrate the peaks associated with C 1s, O 1s in GO and C 1s, O 1s, N 1s, Ag 3d in the hybrid nanocomposite. The intensity of the peak related to C 1s is increased in GO-Ag HN, while the intensity of peak O 1s is reduced, indicating the reduction of GO. The C/O ratio of GO-Ag HN increases twice compared to that of GO due to the removal of oxygen functionalities. Ag 3d peaks in survey spectra provide evidence for the successful anchoring of Ag NPs on GO nanosheets. Figure 4B presents high-resolution C 1s XPS spectra of GO, which show four peaks at 284.7 eV (C=C/C–C in aromatic ring), 286.9 eV (C–OH), 287.3 eV (C=O), and 288.7 eV (O=C–OH).45,49-51 These peaks indicate the presence of different oxygen-containing functional groups (epoxy, hydroxyl, carbonyl, and carboxyl) on the surface of GO nanosheets. High-resolution C 1s XPS spectra of GO-Ag HN are shown in Figure 4C. The considerable increase of the integrated area of sp2 carbon peak is accompanied by the decrease of integrated areas for oxygenated carbon moieties (C–OH and O=C–OH peaks), revealing that epoxy and carboxyl groups help in the anchoring of Ag NPs on GO nanosheets and restoration of sp2 carbon network due to the reduction.
 |
Figure 4 XPS spectra of the GO and GO–Ag HN: (A) survey scans, (B) C 1s of GO, (C) C 1s of GO–Ag HN and (D) Ag 3d from GO-Ag HN. |
Figure 4C shows the high-resolution the Ag 3d spectrum for the GO–Ag HN. There are two peaks of Ag 3d5/2 and Ag 3d3/2 shown in Figure 4D that are centered at binding energies of 367.4 eV and 373.4 eV, respectively, which prove that silver is present only in metallic form, indicating the formation of Ag NPs on the surface of GO nanosheets.51,52
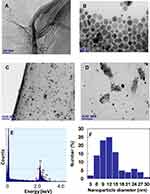
Microscopy investigations (TEM, SEM, AFM) were used to analyze morphological aspects of the GO-Ag HN. TEM image confirms that the thickness of the flat GO nanosheet corresponds to a single atomic layer (Figure 5A). Microscopic wrinkles, curls, and corrugations due to the flexibility of the GO sheets are visible.
During chemical reduction of AgNO3 with PVP, polydispersed Ag NPs were synthesized almost spherical in shape in the size range of 10–40 nm (Figure 5B). Some triangular formations and rods can be also observed. In the case of the hybrid nanocomposite, the positively charged Ag NPs are irregularly precipitated on the negatively charged GO nanosheet surface with almost no aggregation (Figure 5C and D). It was noticed that the amount of Ag NPs is significantly higher in the GO sheet wrinkles, curls, and corrugations (Figure 5D), and at the edges (Figure 5C). Almost no free Ag NPs were observed outside the GO sheets, which demonstrates high interfacial interaction between the Ag NPs and GO. EDX analysis confirmed the presence of Ag NPs on the surface of the GO nanosheet – strong Ag peaks are visible in Figure 5E. The histogram, presented in Figure 5F, indicates that there are variations in the particle sizes ranging from 3 nm to 30 nm, with almost 50% of the particles in the 9–12 nm range.
AFM analysis performed on the thin hybrid nanocomposite film on the silicon surface also reveals Ag NPs embedded into GO nanosheets (Figure 6).
 |
Figure 6 Two-dimensional (A), three-dimensional (B) AFM images and height profile (C) of GO-Ag HN. |
Our previous AFM investigation showed that GO sheet thickness was found to be ca. 8 nm.49 Figure 6A reveals a predominantly hill-valley-structured GO-Ag HN film surface of nanoscale topography. Ag NPs and some of their aggregates are distributed randomly and attached to GO sheets in the agreement with TEM data. The 3D image reveals a pyramid-like morphology of Ag NP aggregates with a height of 5–15 nm and width of about <100 nm (Figure 6B).
Figure 7 shows SEM micrographs of the GO films before and after modification with Ag NPs assembled on silicon surface by vertical dip-coating. Several layers of aggregated and crumpled nanosheets that are closely associated with each other form a GO film with microscopic wrinkles and curls (Figure 7A).
 |
Figure 7 SEM images of GO nanosheet (A) and GO-Ag HN (B–D). |
In the case of the GO-Ag HN (Figure 7B), Ag NPs are deposited on the wrinkled and curled GO sheet surface as supported by TEM and AFM analysis. Detailed SEM analysis reveals that the GO nanosheets undergo partial fragmentation due to the PVP reduction and intensive mixing during the preparation of hybrid nanocomposite. Therefore, visible changes in GO-Ag HN morphology are observed. As can be seen in Figure 7C and D, GO-Ag HN consists of individual closely associated GO nanosheets of leaf-like morphology with sharp edges decorated with Ag NPs. Changes in the lateral size of GO sheets are found to influence antibacterial activities of GO by altering its adsorption abilities, dispersibility, number of corners, and sharp edges.25,53
Pseudomonas aeruginosa Susceptibility to Antibiotics
Growing antibiotic resistance is complicating the treatment of patients and increasing the risk of fatal outcomes. Nosocomial infections caused by P. aeruginosa are a major threat in the hospital environment, since MDR strains are already proven to carry a higher risk of severe illness.3 Antibiotic resistance was determined for all 64 clinical strains of P. aeruginosa. The resistance patterns differed from those seen in European hospitals in a 2011–12 study by Sader et al.54 Most of the strains were resistant to ciprofloxacin – 79.7% (n=51), meropenem – 76.6% (n=49), gentamicin – 71.9% (n=46), and piperacillin – 56.3% (n=36). Almost half of the tested strains were resistant to ceftazidime (43.5%, n=28). The most effective antibiotic for the tested strains was amikacin. Only 28.1% (n=18) of strains were resistant to this antibiotic.
Molecular testing showed that the most prevalent antibiotic resistance gene among P. aeruginosa strains was the AMPC beta-lactamase gene (65.6%, n=42). Prevalence of the VIM gene was higher than described by Castanheira et al.55 The VIM gene was found in 43.8% (n=28) of isolated P. aeruginosa strains, while ESBL genes were found in 4.7% (n=3) of strains with one strain having PER gene and two strains having GES gene. The presence of two resistance genes was detected for 17.2% (n=11) of P. aeruginosa strains, with a combination on AMPC and VIM accounting for all of the cases.
When results of antibiotic resistance testing were matched to resistance gene presence, no clear relation was found among strains resistant to meropenem and piperacillin (Table 1). More of the strains resistant to meropenem were negative for AmpC and ESBL genes (90.1% (n=20) and 78.7% (n=48), respectively). Among meropenem-resistant strains, 69.0% (n=29) were positive for AmpC and 33.3% (n=1) for ESBL genes. In contrast, among strains positive for MBL genes, more were resistant to meropenem (82.1%, n=23) compared to MBL negative ones (72.2% (n=26)). Regardless of the presence of the AmpC, ESBL, and MBL genes from 50% to 67% of all tested P. aeruginosa strains were resistant to piperacillin.
 |
Table 1 Antibiotic Susceptibility of Resistant P. aeruginosa Grouped by Presented Resistance Genes |
P. aeruginosa strains that tested negative for the AmpC gene were significantly more resistant to cefepime than positive ones (77.3% (n=17) and 26.2% (n=11), respectively, p<0.001). ESBL genes were present in all of cefepime-resistant strains (100% (n=3)), while 41.0% (n=25) of ESBL negative were resistant. A similar pattern was observed when comparing AmpC negative and positive P. aeruginosa strains in regard to ciprofloxacin resistance. Of strains negative for the AmpC gene, 95.5% (n=21) were resistant to ciprofloxacin, while among positive ones only 66.7% (n=28) were resistant (p=0.045). ESBL negative strains were ciprofloxacin-resistant in 82.0% (n=50) of cases, while positive ones were resistant in 66.6% (n=2) of cases. Of MBL gene positive P. aeruginosa strains, 89.3% (n=25) were ciprofloxacin-resistant, while among MBL negative strains 75.0% (n=27) were resistant.
AmpC negative P. aeruginosa strains were resistant to gentamicin in significantly higher rates than positive strains (95.5% (n=21) and 66.7% (n= 28), respectively, p=0.012). Strains positive for MBL genes were significantly more resistant to gentamicin than strains without these genes (92.9% (n=26) and 63.9% (n=23), respectively, p=0.008). Amikacin resistance was strongly associated with genes that determine beta-lactamatic activity determining genes. Strains that tested negative for AmpC were more resistant to amikacin than positive ones (72.7% (n=16) and 4.8% (n=16), respectively, p<0.001). All of the ESBL producing strains (100.0% (n=3)) were amikacin resistant, while only 24.6% (n=15), p=0.02 ESBL negative strains showed resistance. More than half (53.6% (n=15)) of MBL producing strains were resistant to amikacin, while only 8.3% (n=3) of the MBL negative strains were resistant.
Antibacterial Effect of the GO-Ag HN on MDR P. aeruginosa
Our results followed previously revealed trends of GO-metal nanoparticle based composites being effective bactericidal agents against a wide range of bacteria strains due to the possible synergy of the multiple toxicity mechanisms.49 In this study, the investigations were performed to elucidate the toxicity mechanisms of GO-Ag HN on P. aeruginosa strains with acquired resistance genes. The effect of the hybrid GO-Ag HN on the bacterial viability was tested on all 64 isolated clinical strains of P. aeruginosa. A clear correlation between growth intensity and incubation time (up to 10 min) before plating was observed (p<0.001). Most of the strains (78.1%, n=50) showed abundant growth when plated without incubation, while 20.3% (n=13) of strains showed sparse or average growth, and only one strain was completely inhibited. Only two strains remained growing abundantly after 5 min of incubation. The breakpoint where total growth inhibition was the most common outcome appeared after 6 min of incubation, when the growth of 43.8% (n=28) of P. aeruginosa strains was not observed. The number of strains that were inhibited decreased after long time incubation comparing to the 10-minute mark (Figure 8).
 |
Figure 8 The growth intensity distribution of GO-Ag hybrid nanocomposite effected P. aeruginosa. The graph shows rapid decline of P. aeruginosa growth in first 6 minutes of incubation. |
No clear relation was found between growth intensity and resistance to amikacin, ceftazidime, ciprofloxacin, and piperacillin. When resistance to meropenem was taken into account, a difference of growth intensity appeared between susceptible and resistant strains. Meropenem-resistant strains were more viable than susceptible ones at incubation time from 0 to 10 min (Table 2). Despite the fact that statistically significant difference was achieved only at incubation times of 1, 4, and 10 min (p=0.036, p=0.034, and p=0.039, respectively), the greater viability of resistant strains was seen in the incubation time range of 0–10 min. After 2 h incubation, an interesting trend appeared: contrary to previous results a greater proportion of meropenem susceptible strains showed growth than meropenem-resistant ones. This trend was even stronger in the case of gentamicin-resistant strains: after 2 h incubation, susceptible strains were significantly more likely to grow than resistant ones. When considering the total number of colonies and meropenem resistance level, a statistically significant correlation was found on incubation times of 1–10 min.
 |
Table 2 The Dispersion of P. aeruginosa Growth Intensity Dependant on Meropenem Susceptibility Status |
Molecular testing showed that only three P. aeruginosa strains had ESBL genes, while 28 strains had the VIM gene. When strains were affected by GO-Ag HN, the presence of ESLB genes was associated with lower viability in the case when no incubation was performed before plating (p<0.001). Completely opposite results were obtained when VIM positive strains were incubated with GO-Ag HN (Figure 9). After 1 min of incubation, growth was significantly more intense among VIM positive strains (p=0.017). However, after 2 h incubation, only VIM negative strains grew, when no colonies were formed by VIM positive strains (p=0.012). No significant difference in growth intensity was observed between strains positive and negative for the AMPC gene.
The structural changes of the bacteria cell membrane after GO-Ag HN treatment were elucidated at different times using AFM analysis. The images of untreated P. aeruginosa show rod-shaped morphology with a length of 2.5–3 μm (Figure 10A). The cell membrane is undamaged; therefore, the profile of the cell depicts an intact and relatively smooth cell surface without ruptures and bulges. Significant changes in the cell morphology of GO-Ag HN treated P. aeruginosa cells were obtained. After incubation for 10 min with GO-Ag HN, P. aeruginosa cells maintained rod-shaped morphology, but some bulges, sags, and pores appear on the surface of cells, as shown in Figure 10B. Besides, some micelle-like structures are also visible on the edges of cells suggesting damage to the P. aeruginosa outer membrane. The unevenness of the surface increased and cells lost their rod-shaped morphology and the borders of cells were diffused after 30 min incubation with GO-Ag HN (Figure 10C). Such changes reveal membrane damage that increases permeability and membrane leakage and results in cell lysis.
The height profile analysis provided a quantitative evaluation of bacteria dimension changes. As seen in Figure 10D, bacteria height decreases significantly as the duration of incubation with GO-Ag HN increases. After incubation for 120 min, P. aeruginosa height becomes close to that of free GO-Ag HN film (see Figure 6), and only the cell debris was noted, indicating bacterial death.
Discussion
A Gram-negative opportunistic pathogen, with active movement and obligate oxygen demand, Pseudomonas aeruginosa is one of the major pathogenic bacteria in health-care settings, and can lead to chronic infections and acquire antibiotic tolerance. Nosocomial infections caused by P. aeruginosa are a major threat in a hospital environment, since MDR strains are already proven to carry a higher risk of severe illness. Growing antibiotic resistance is complicating patient treatment and increasing the risk of fatal outcome.3
At the present time, prevention and therapy of P. aeruginosa infections has become increasingly challenging, owing to its intrinsic and acquired drug-resistance properties. Carbapenems have been the most important therapeutic option when dealing with multidrug-resistant P. aeruginosa for decades.56 In this study, we isolated and tested 64 imipenem-resistant clinical P. aeruginosa strains. Among them, 59 were multidrug-resistant. The most concerning result was that meropenem resistance was found in 49 of the strains (more than 76%). The threat posed by antibiotic-resistant P. aeruginosa strains requires great efforts to develop highly effective and safe bactericidal products with a wide spectrum of activity.57
In recent years, there has been increased interest in GO-based nanocomposites for use to control multidrug-resistant pathogens such as P. aeruginosa. The incorporation of inorganic nanostructures on the GO surface has enhanced the antibacterial efficiency of the nanocomposite. The most effective inorganic nanostructures against bacterial infections are Ag nanoparticles. They can be prepared by various techniques. In this study, we synthesized Ag NPs by polyol reduction method in the presence of PVP. PVP was used not only as a reducing agent but also as a protection of the Ag NPs from growing and agglomerating due to the steric effect. We chose PVP of relatively high molecular weight (Mw = 40,000 mol−1), because longer chains cause a higher steric effect.46 After the introduction of PVP, Ag+ ions or Ag0 particles interact with N or O in PVP, and a covered layer was generated on the surface. Zhang et al58 analyzed nanoparticles with a diameter of about 50 nm and assumed that interactions with N and O were equal. However, Wang et al59 proved that in the case of Ag nanoparticles with a diameter smaller than 50 nm, the coordination between N and silver was the main reaction, while interaction with O was less important. Thus, the main mechanism for protection of Ag nanoparticles by PVP was N, in PVP, coordinating with silver and forming a protection layer as presented in Figure 1. In addition, we found that the dispersion of PVP-stabilized silver nanoparticles has considerably increased stability. Tejamaya et al60 have shown that dispersion of PVP-stabilized Ag NPs was stable over 21 days with only small losses in total concentration; also, no changes of shape, aggregation, or dissolution compared to other synthesis methods were observed. Our UV-vis and AFM investigations showed that PVP-stabilized Ag NPs, nearly spherical in shape, with a mean size of 25 nm were synthesized. However, after precipitation on the surface of GO sheets, the main size of Ag NPs decreased to 9–12 nm with almost no aggregation (Figure 5C, D, and F), because GO prevented nanoparticles from aggregation better than PVP.61 Ag NPs of such size are able to attach to the surface of the bacteria and even penetrate the cell wall.20
We found49 that GO nanosheets show toxicity on different Gram-positive and Gram-negative bacterial strains, including microencapsulation ability. The GO antibacterial mechanism can be related to the mechanical destruction of membranes, redox reaction with biomolecules, and catalysis of extracellular metabolites.25,29 As can be seen in Figure 7C and D, GO undergoes morphological changes after addition of PVP-stabilized Ag NPs, and leaf-like GO nanosheets of the size ranging from ca. 300 nm to 600 nm with sharp edges were formed. Perreault et al53 found that GO-based surface coatings showed higher antimicrobial activity in smaller GO sheet sizes, which could be ascribed to the higher defect density. Sharp edges and corners of the sheets can cause physical damage to the cell membrane upon direct contact with bacteria.29 GO-based nanostructures began to show an antibacterial effect when the contact angle was 37°, reached a maximum at 90°,62 and may result in the formation of pores in bacterial cell walls, osmotic imbalance, and cell death. SEM analysis of GO-Ag HN showed that the edges of the fragmented GO nanosheets are densely decorated with Ag NPs (Figure 7D). This can enhance the penetration capacity of Ag NPs after the sharp edges of GO spontaneously pierce and cause cell membrane defects. Discussions on the mechanisms of Ag NP interaction with bacterial cells are continuing despite a remarkable amount of experimental data on their toxicity and mode of action.15,18,21,22 Dakal et al21 described that Ag NPs can interact with Gram-negative bacteria in several ways: 1) the electrostatic interaction between positively charged Ag NPs and the negatively charged cell membrane facilitates Ag NP attachment onto the cell membrane. Morphological changes are caused by shrinkage of the cytoplasm and membrane detachment, finally leading to the rupture of the cell wall. 2) Ag NPs can penetrate the bacteria and interact with sulfur- and phosphorus-containing structures, such as proteins, lipids, and DNA, resulting in their deactivation. 3) The increased concentration of Ag+ ions is expected to generate free radicals and reactive oxygen species (ROS) and a consequent increase in cellular oxidative stress that promotes both cytotoxic as well as genotoxic effects. 4) The Ag NP effect on bacterial signal transduction pathways that affect bacterial growth and other molecular and cellular activities.
We obtained significant changes in the cell morphology of P. aeruginosa cells after incubation for 10 min with GO-Ag HN. Some bulges, pores, and micelle-like structures appear on the surface of a cell, suggesting damage to the outer membrane. Different metal nanoparticles appear to have a similar effect on bacteria cells, eg, when bacteria were treated with Fe3O4, NP roughness was reduced several times compared to untreated samples.63
In this study, we showed that GO-Ag HN reduces P. aeruginosa bacterial strain survival and ability to form colonies. This potentially happens due to a higher probability of bacterial cells colliding with GO-Ag HN particles in a liquid medium. Our synthesized GO-Ag HN appears to have a much faster effect. It took about 7 minutes to inhibit the growth of most P. aeruginosa strains instead of 2 to 3 hours as was described by Prasad et al.64 The higher survival rate of bacterial cells was noticed among meropenem-resistant P. aeruginosa strains. This might be associated with metallo beta-lactamase presence, since P. aeruginosa strains testing positive for these genes were more likely to be meropenem resistant and form more bacterial colonies after incubation with GO-Ag HN. It is possible that enzymes encoded by metallo beta-lactamase genes help to protect bacterial cells from damage or help to repair them afterward.
Conclusion
A simple two-stage methodology has been performed for the synthesis of polyvinylpyrrolidone-stabilized silver and graphene oxide hybrid nanocomposite (GO-Ag HN). GO nanosheets were decorated with spherical Ag NPs with a main diameter of 9–12 nm. During preparation GO nanosheets undergo partial fragmentation and coatings are composed of closely associated 300–600 nm sheets with an increased amount of sharp edges positioned at various angles. This GO-Ag HN possesses enhanced contact-based antibacterial activity against antibiotic-resistant bacteria strains, such as P. aeruginosa strains with acquired resistance genes. An incubation time of only 7 min was enough to inhibit the growth of the most isolated clinical P. aeruginosa strains. Interestingly, meropenem-resistant strains showed higher tolerance to GO-Ag HN, which is associated with biochemical or morphological properties of these strains. After GO-Ag HN acts on P. aeruginosa, the cell membrane becomes shriveled, damaged and constituents leak out. These results indicate that GO-Ag HN is a promising antibacterial agent to control nosocomial infections caused by antibiotic-resistant bacteria strains.
Acknowledgments
A cooperative project between Kaunas University of Technology (KTU) and Lithuanian University of Health Sciences (LSMU) No. PP 34/146.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Exner M, Bhattacharya S, Christiansen B, et al. Antibiotic resistance: what is so special about multidrug-resistant Gram-negative bacteria? GMS Hyg Infect Control. 2017;12:Doc05.
2. Baptista PV, McCusker MP, Carvalho A, et al. Nano-strategies to fight multidrug resistant bacteria – “A battle of the titans”. Front Microbiol. 2018;9:1441. doi:10.3389/fmicb.2018.01441
3. Micek ST, Wunderink RG, Kollef MH, et al. An international multicenter retrospective study of Pseudomonas aeruginosa nosocomial pneumonia: impact of multidrug resistance. Crit Care. 2015;19(1):219. doi:10.1186/s13054-015-0926-5
4. Breidenstein EB, de la Fuente-núñez C, Hancock RE. Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol. 2011;19(8):419–426. doi:10.1016/j.tim.2011.04.005
5. Croughs PD, Klaassen CHW, van Rosmalen J, et al. Unexpected mechanisms of resistance in Dutch Pseudomonas aeruginosa isolates collected during 14 years of surveillance. Int J Antimicrob Agents. 2018;52(3):407–410. doi:10.1016/j.ijantimicag.2018.05.009
6. Potron A, Poirel L, Nordmann P. Emerging broad-spectrum resistance in Pseudomonas aeruginosa and Acinetobacter baumannii: mechanisms and epidemiology. Int J Antimicrob Agents. 2015;45(6):568–585. doi:10.1016/j.ijantimicag.2015.03.001
7. Amini A, Namvar AE. Antimicrobial resistance pattern and presence of beta-lactamase genes in Pseudomonas aeruginosa strains isolated from hospitalized patients, Babol-Iran. J Med Bacteriol. 2019;8(1–2):45–50.
8. Levy SB. Factors impacting on the problem of antibiotic resistance. J Antimicrob Chemother. 2002;49(1):25–30. doi:10.1093/jac/49.1.25
9. Judd WR, Ratliff PD, Hickson RP, et al. Clinical and economic impact of meropenem resistance in Pseudomonas aeruginosa–infected patients. Am J Infect Control. 2016;44(11):1275–1279. doi:10.1016/j.ajic.2016.04.218
10. Hemeg HA. Nanomaterials for alternative antibacterial therapy. Int J Nanomed. 2017;12:8211–8225. doi:10.2147/IJN.S132163
11. Wang L, Hu C, Shao L. The antimicrobial activity of nanoparticles: present situation and prospects for the future. Int J Nanomed. 2017;12:1227–1249. doi:10.2147/IJN.S121956
12. Natan M, Banin E. From nano to micro: using nanotechnology to combat microorganisms and their multidrug resistance. FEMS Microbiol Rev. 2017;41(3):302–322. doi:10.1093/femsre/fux003
13. Mehran A, Mahendra R. Recent advances in antibacterial applications of metal nanoparticles (MNPs) and metal nanocomposites (MNCs) against multidrug-resistant (MDR) bacteria. Expert Rev Anti Infect. 2019;17(6):419–428. doi:10.1080/14787210.2019.1614914
14. Pelgrift RY, Friedman AJ. Nanotechnology as a therapeutic tool to combat microbial resistance. Adv Drug Deliver Rev. 2013;65(13–14):1803–1815. doi:10.1016/j.addr.2013.07.011
15. Gold K, Slay B, Knackstedt M, Gaharwar AK. Antimicrobial activity of metal and metal-oxide based nanoparticles. Adv Ther. 2018;1(3):1700033. doi:10.1002/adtp.201700033
16. Güzel R, Erdal G. Synthesis of silver nanoparticles. In: Maaz K, editor. Silver Nanoparticles - Fabrication, Characterization and Applications. IntechOpen; 2018:1–20.
17. Singh R, Shedbalkar UU, Wadhwani SA, Chopade BA. Bacteriagenic silver nanoparticles: synthesis, mechanism, and applications. Appl Microbiol Biotechnol. 2015;99(11):4579–4593. doi:10.1007/s00253-015-6622-1
18. Salomoni R, Léo P, Montemor AF, Rinaldi BG, Rodrigues MFA. Antibacterial effect of silver nanoparticles in Pseudomonas aeruginosa. Nanotechnol Sci Appl. 2017;10:115–121. doi:10.2147/NSA.S133415
19. Barros CHN, Fulaz S, Stanisic D, Tasic L. Biogenic nanosilver against multidrug-resistant bacteria (MDRB). Antibiotics. 2018;7(3):
20. Liao S, Zhang Y, Pan X, et al. Antibacterial activity and mechanism of silver nanoparticles against multidrug-resistant Pseudomonas aeruginosa. Int J Nanomed. 2019;14:1469–1487. doi:10.2147/IJN.S191340
21. Dakal T, Kumar A, Majumdar RS, Yadav V. Mechanistic basis of antimicrobial actions of silver nanoparticles. Front Microbiol. 2016;7:1831. doi:10.3389/fmicb.2016.01831
22. Durán N, Durán M, de Jesus MB, Seabra AB, Fávaro WJ, Nakazato G. Silver nanoparticles: a new view on mechanistic aspects on antimicrobial activity. Nanomed-Nanotechnol. 2016;12(3):789–799. doi:10.1016/j.nano.2015.11.016
23. Kedziora A, Speruda M, Krzyzewska E, Rybka J, Lukowiak A, Bugla-Płosko G. Similarities and differences between silver ions and silver in nanoforms as antibacterial agents. Int J Mol Sci. 2018;19(2):
24. Tegou E, Magana M, Katsogridaki AE, et al. Terms of endearment: bacteria meet graphene nanosurfaces. Biomaterials. 2016;89:38–55. doi:10.1016/j.biomaterials.2016.02.030
25. Zheng H, Ma R, Gao M, et al. Antibacterial applications of graphene oxides: structure-activity relationships, molecular initiating events and biosafety. Sci Bull. 2018;63:133–142. doi:10.1016/j.scib.2017.12.012
26. Lukowiak A, Kedziora A, Strek W. Antimicrobial graphene family materials: progress, advances, hopes and fears. Adv Colloid Interface. 2016;236:101–112. doi:10.1016/j.cis.2016.08.002
27. Wu X, Tan S, Xing Y, Pub Q, Wub M, Zhaoa JX. Graphene oxide as an efficient antimicrobial nanomaterial for eradicating multi-drug resistant bacteria in vitro and in vivo. Colloids Surf B. 2017;157:1–9. doi:10.1016/j.colsurfb.2017.05.024
28. Nanda SS, Yi DK, Kim K. Study of antibacterial mechanism of graphene oxide using Raman spectroscopy. Sci Rep. 2016;6:28443. doi:10.1038/srep28443
29. Xia M-Y, Xie Y, Yu C-H, et al. Graphene-based nanomaterials: the promising active agents for antibiotics-independent antibacterial applications. J Control Release. 2019;307:16–31. doi:10.1016/j.jconrel.2019.06.011
30. Akhavan O, Ghaderi E. Toxicity of graphene and graphene oxide nanowalls against bacteria. ACS Nano. 2010;4:5731–5736. doi:10.1021/nn101390x
31. Romero-Vargas Castrillón S, Perreault F, de Faria AF, Elimelech M. Interaction of graphene oxide with bacterial cell membranes: insights from force spectroscopy. Environ Sci Technol Lett. 2016;2:112–117. doi:10.1021/acs.estlett.5b00066
32. Yousefi M, Dadashpour M, Hejazi M, et al. Anti-bacterial activity of graphene oxide as a new weapon nanomaterial to combat multidrug-resistance bacteria. Mater Sci Eng C. 2017;74:568–581. doi:10.1016/j.msec.2016.12.125
33. Muszynski R, Seger B, Kamat PV. Decorating graphene sheets with gold nanoparticle. J Phys Chem A. 2008;112(14):5263–5266.
34. Ji H, Sun H, Qu X. Antibacterial applications of graphene-based nanomaterials: recent achievements and challenges. Adv Drug Deliv Rev. 2016;105:176–189. doi:10.1016/j.addr.2016.04.009
35. de Moraes ACM, Lima BA, de Faria AF, Brocchi M, Alves OL. Graphene oxide-silver nanocomposite as a promising biocidal agent against methicillin-resistant Staphylococcus aureus. Int J Nanomed. 2015;10:6847–6861. doi:10.2147/IJN.S90660
36. Zhu Z, Su M, Ma L, Ma L, Liu D, Wang Z. Preparation of graphene oxide-silver nanoparticle nanohybrids with highly antibacterial capability. Talanta. 2013;117:449–455. doi:10.1016/j.talanta.2013.09.017
37. Mikucionyte G, Zamorano L, Vitkauskiene A, et al. Nosocomial dissemination of VIM-2-producing ST235 Pseudomonas aeruginosa in Lithuania. Eur J Clin Microbiol Infect Dis. 2016;35(2):195–200. doi:10.1007/s10096-015-2529-0
38. Jankauskaitė V, Lozovskis P, Valeika V, Vitkauskienė A Graphene oxide and metal particles nanocomposites for inhibition of pathogenic bacteria strains.
39. Oldenburg SJ Silver nanoparticles: properties and applications. Available from: https://www.sigmaaldrich.com/technical-documents/articles/materials-science/nanomaterials/silver-nanoparticles.html.
40. Knauer A, Thete A, Li S, et al. Au/Ag/Au double shell nanoparticles with narrow size distribution obtained by continuous micro segmented flow synthesis. Chem Eng J. 2011;166(3):1164–1169. doi:10.1016/j.cej.2010.12.028
41. Çiplak Z, Yildiz N, Çalimli A. Investigation of graphene/Ag nanocomposites synthesis parameters for two different synthesis methods. Fuller Nanotub Car N. 2014;23(4):361–370. doi:10.1080/1536383X.2014.894025
42. Gurunathan S, Han JW, Park JH, et al. Reduced graphene oxide–silver nanoparticle nanocomposite: a potential anticancer nanotherapy. Int J Nanomed. 2015;10:6257–6276. doi:10.2147/IJN.S92449
43. He D, Peng Z, Gong W, Luo Y, Zhao P, Kong L. Mechanism of a green graphene oxide reduction with reusable potassium carbonate. RSC Adv. 2015;5:11966–11972. doi:10.1039/C4RA14511A
44. Sharma N, Sharma V, Jain Y, et al. Synthesis and characterization of graphene oxide (GO) and reduced graphene oxide (rGO) for gas sensing application. Macromol Symp. 2017;376:1700006. doi:10.1002/masy.201700006
45. Shao W, Liu X, Min H, et al. Preparation, characterization, and antibacterial activity of silver nanoparticle-decorated graphene oxide nanocomposite. ACS Appl Mater Interfaces. 2015;7:6966–6973. doi:10.1021/acsami.5b00937
46. Bryaskova R, Pencheva D, Nikolov S, Kantardjiev T. Synthesis and comparative study on the antimicrobial activity of hybrid materials based on silver nanoparticles (AgNps) stabilized by polyvinylpyrrolidone (PVP). J Chem Biol. 2011;4:185–191. doi:10.1007/s12154-011-0063-9
47. Chen S, Cheng B, Ding C. Synthesis and characterization of poly(vinyl pyrrolidone)/reduced Graphene oxide nanocomposite. J Macromol Sci B Phys. 2015;54:481–491. doi:10.1080/00222348.2015.1010433
48. Song Y-J, Wang M, Zhang X-Y, Wu J-Y, Zhang T. Investigation on the role of the molecular weight of polyvinyl pyrrolidone in the shape control of high-yield silver nanospheres and nanowires. Nanoscale Res Lett. 2014;9(1):17. doi:10.1186/1556-276X-9-17
49. Jankauskaitė V, Vitkauskienė A, Lazauskas A, Baltrusaitis J, Prosyčevas I, Andrulevičius M. Bactericidal effect of graphene oxide/Cu/Ag nanoderivatives against Escherichia coli, Pseudomonas aeruginosa, Klebsiella pneumoniae, Staphylococcus aureus and Methicillin-resistant Staphylococcus aureus. Int J Pharm. 2016;511:90–97. doi:10.1016/j.ijpharm.2016.06.121
50. Jia Z, Li C, Liu D, Jiang L. Direct hydrothermal reduction of graphene oxide based papers obtained from tape casting for supercapacitor applications. RSC Adv. 2015;5:81030–81037. doi:10.1039/C5RA17277B
51. Shaikh A, Parida S, Bohmb S. One step eco-friendly synthesis of Ag–reduced graphene oxide nanocomposite by phytoreduction for sensitive nitrite determination. RSC Adv. 2016;6:100383–100391. doi:10.1039/C6RA23655C
52. Khan ME, Khan MM, Cho MH. Biogenic synthesis of a Ag–graphene nanocomposite with efficient photocatalytic degradation, electrical conductivity and photoelectrochemical performance. N J Chem. 2015;39:8121–8129. doi:10.1039/C5NJ01320H
53. Perreault F, de Faria AF, Nejati S, Elimelech M. Antimicrobial properties of graphene oxide nanosheets: why size matters. ACS Nano. 2018;9(7):7226–7236. doi:10.1021/acsnano.5b02067
54. Sader HS, Farrell DJ, Castanheira M, Flamm RK, Jones RN. Antimicrobial activity of ceftolozane/tazobactam tested against Pseudomonas aeruginosa and Enterobacteriaceae with various resistance patterns isolated in European hospitals (2011–12). J Antimicrob Chemother. 2014;69(10):2713–2722. doi:10.1093/jac/dku184
55. Castanheira M, Deshpande LM, Costello A, Davies TA, Jones RN. Epidemiology and carbapenem resistance mechanisms of carbapenem-non-susceptible Pseudomonas aeruginosa collected during 2009–11 in 14 European and Mediterranean countries. J Antimicrob Chemother. 2014;69(7):1804–1814. doi:10.1093/jac/dku048
56. Livermore D, Woodford N. Carbapenemases: a problem in waiting? Curr Opin Microbiol. 2000;3(5):489–495. doi:10.1016/S1369-5274(00)00128-4
57. Yayan J, Ghebremedhin B, Rasche K, Webber MA. Antibiotic resistance of Pseudomonas aeruginosa in pneumonia at a Single University Hospital Center in Germany over a 10-year period. PLoS One. 2015;10(10):e0139836. doi:10.1371/journal.pone.0139836
58. Zhang Z, Zhao B, Hu L, Protective PVP. PVP protective mechanism of ultrafine silver powder synthesized by chemical reduction processes. J Solid State Chem. 1996;121(1):105–110. doi:10.1006/jssc.1996.0015
59. Wang H, Qiao X, Chen J, Wang X, Ding S. Mechanisms of PVP in the preparation of silver nanoparticles. Mater Chem Phys. 2005;94:449–453. doi:10.1016/j.matchemphys.2005.05.005
60. Tejamaya M, Römer I, Merrifield RC, Lead JR. Stability of citrate, PVP, and PEG coated silver nanoparticles in ecotoxicology media. Environ Sci Technol. 2012;46:7011–7017. doi:10.1021/es2038596
61. Dat NM, Linh VNP, Huy LA, et al. Fabrication and antibacterial activity against Pseudomonas aeruginosa and Staphylococcus aureus of silver nanoparticle decorated reduced graphene oxide nanocomposites. Mater Technol. 2019;34(7):369–375. doi:10.1080/10667857.2019.1575555
62. Pham VT, Truong VK, Quinn MD, et al. Graphene induces formation of pores that kill spherical and rod-shaped bacteria. ACS Nano. 2015;9(8):8458–8467. doi:10.1021/acsnano.5b03368
63. Alavi M, Karimi N. Ultrasound assisted-phytofabricated Fe3O4 NPs with antioxidant properties and antibacterial effects on growth, biofilm formation, and spreading ability of multidrug resistant bacteria. Artif Cells Nanomed Biotechnol. 2019;47(1):2405–2423. doi:10.1080/21691401.2019.1624560
64. Prasad K, Lekshmi G, Ostrikov S, et al. Synergic bactericidal effects of reduced graphene oxide and silver nanoparticles against Gram-positive and Gram-negative bacteria. Sci Rep. 2017;7:1591. doi:10.1038/s41598-017-01669-5
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.