Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 15
DNTTIP2 Expression is Associated with Macrophage Infiltration and Malignant Characteristics in Low-Grade Glioma
Authors Liu YJ , Zeng SH, Qian WH, Tao MX, Zhu YY, Li JP
Received 12 January 2022
Accepted for publication 4 March 2022
Published 24 March 2022 Volume 2022:15 Pages 261—275
DOI https://doi.org/10.2147/PGPM.S356326
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin H Bluth
Yuan-Jie Liu,1,2,* Shu-hong Zeng,2,3,* Wei-hua Qian,1 Min-xian Tao,1 Ying-ying Zhu,4 Jie-pin Li1– 3
1Department of Oncology, Zhangjiagang TCM Hospital Affiliated to Nanjing University of Chinese Medicine, Zhangjiagang, Jiangsu, 215600, People’s Republic of China; 2Department of Oncology, Affiliated Hospital of Nanjing University of Chinese Medicine, Jiangsu Province Hospital of Chinese Medicine, Nanjing, Jiangsu, 210029, People’s Republic of China; 3No. 1 Clinical Medical College, Nanjing University of Chinese Medicine, Nanjing, Jiangsu, 210023, People’s Republic of China; 4Department of Oncology, Affiliated Aoyang Hospital of Jiangsu University, Zhangjiagang, Jiangsu, 215600, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jie-pin Li, Email [email protected]
Purpose: Low-grade gliomas (LGG) are primary brain tumors that often affect predominantly young adults, which usually have a painless course, and have a longer survival period compared to patients with high-grade gliomas. Relatively established treatment options include surgery, radiotherapy, chemotherapy or combination therapy, as well as individualized management based on tumor location, histology, molecular features and patient characteristics. Due to the rapid development of targeted therapies, the development of new molecular targets is now a very promising research direction.
Methods: We explored the diagnostic value, clinical relevance, and molecular function of deoxynucleotidyl transferase terminal-interacting proteins 2 (DNTTIP2) in LGG using MethSurv, MEXPRESS, STRING, cBioPortal, Tumor Immunity Estimation Resource (TIMER) database, Gene Expression Profiling Interactive Analysis (GEPIA) databases. Besides, the “CIBERSORT” algorithm was conducted to estimate immune cells infiltration abundance, with “ggplot2” package visualizing the results. In vivo and vitro experiments were used to verify the speculations of bioinformatics analysis.
Results: In LGG patients, DNTTIP1/2 were over-expressed at mRNA levels and high DNTTIP1/2 levels correlated with poor survival in LGG patients. We confirmed that DNTTIP2 significantly promotes M2 macrophage activation and angiogenesis, which may be related to the IL6/JAK/STAT3 signaling pathway. In addition, we found that DNTTIP2 amplification was associated with an unfavorable prognosis in LGG patients. We demonstrated, finally, a correlation between DNTTIP2 gene hypermethylation and a poor prognosis in LGG.
Conclusion: This study demonstrated that DNTTIP1/2 had diagnostic and prognostic value in LGG patients. The biological mechanisms of DNTTIP2 regarding angiogenesis and macrophage activation may provide new insights into the treatment of glioma.
Keywords: low-grade glioma, DNTTIP2, angiogenesis, M2 macrophages infiltration, bioinformatics
Introduction
Glioma is the most frequent central nervous system malignancy.1 The World Health Organization (WHO) classification divides glioma into four grades (grades I–IV).2 Low-grade gliomas (LGGs) are defined as diffuse low- and moderate-grade gliomas (WHO grades II and III). Because of its high malignancy and high degree of recurrence, LGG may rapidly progress to glioblastoma (GBM).3 Despite a significant amount of effort devoted to LGG research, specifics of its pathogenesis remain unclear and there have been no breakthroughs made in its treatment, usually surgery followed by post-operative radiotherapy and chemotherapy. Therefore, research into the pathogenesis of glioma and early diagnosis of glioma is particularly important.
With the rise of bioinformatics and high-throughput sequencing technologies, some progress has been made in identifying molecular markers related to LGG progression which can be used to predict prognosis and also a patient’s response to personalized treatment.
Deoxynucleotidyl transferase terminal-interacting proteins 1/2 (DNTTIP1/2) were initially identified as regulators of terminal deoxynucleotidyl transferase (TDT), which plays a role in transcription regulation. DNTTIP1/2 is homologous to the transcription factor p65/NF-κB and acts on the histone deacetylases 1 and 2 (HDAC1 and HDAC2).4,5 Epigenetic changes have been discovered in all stages of cancer progression6 and histone acetylation is one of the most studied epigenetic regulatory pathways. Histone deacetylases (HDACs) are closely related to cancer occurrence and progression, and p53 deacetylation has been reported to be associated with the failure of cancer suppression.7 HDAC1 binds to DNTTIP1 which may be involved in this mechanism.8 DNTTIP1 has been shown to be a pro-oncogenic factor and a likely therapeutic target in non-small cell lung cancer (NSCLC) and oral squamous cell carcinoma (OSCC).9 DNTTIP2 binds to DNA and core histones which contain an acid-rich amino acid region at their C-termini and is thought to act as a histone chaperone in the nucleus. The role of DNTTIP2 in cancer is not known. We hypothesized that DNTTIP2 may be a biomarker for LGG diagnosis and a potential target for anti-LGG therapy.
In this present study, we conducted a bioinformatic analysis and experimental validation were performed to reveal the potential functions of DNTTIP2 in LGG. Our results suggested that DNTTIP2 might be a novel prognostic biomarker and treatment target for LGG.
Materials and Methods
Flowchart of the Study
Figure 1 illustrates the study workflow.
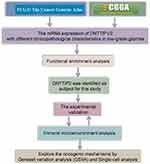 |
Figure 1 Study workflow. |
Reagents
The reagents and antibodies used are listed in the Supplementary material (Supplementary Table 1). The concentrations used were based on those used in previous research or on the manufacturers’ recommendations. The specific experimental details are provided in Supplementary material.
Cell Culture
Human glioma U87MG, U251, SHG-44, H4 cells and Human umbilical vein endothelial cells (HUVEC) were acquired from Procell Life Science &Technology (Wuhan, China) grown in Dulbecco’s Modified Eagle Medium (DMEM) with 10% fetal bovine serum (FBS) in a 5% CO2 (Incubator) at 37°C. All media were supplemented with 1% penicillin/streptomycin. Cells passed regular mycoplasma contamination testing.
Western Blot
The Western blotting method followed previous descriptions.10 Proteins were lysed in RIPA buffer and protein concentrations were measured by the Bradford assay. Samples of 20 µg each were separated on 10% or 8% SDS-PAGE, proteins were electro-transferred to polyvinylidene fluoride (PVDF) membranes, and blocked with 5% bovine serum albumin. The blots were then probed with the relevant primary antibodies at 4 °C overnight. Blots were washed in Tris-buffered saline containing 0.05% Tween-20 and were incubated with the corresponding secondary antibodies with an Electrochemiluminescence (ECL) detection kit used to measure densities. The protein bands (including β-actin as the loading control) were visualized with a gel documentation system (ChemiDoc XRS+) and relative concentrations were determined.
RNAi and Overexpression Plasmid Construction and Transfection
U87MG and U251 cells which express relatively high levels of DNTTIP2 were selected for subsequent experiments. The plasmids described below were all constructed by GeneChem (Shanghai, China). The procedures for constructing over-expression and RNAi plasmids are described in the Supplementary material. Three short hairpins interfering RNAs (shRNA) targeting DNTTIP2 were constructed and the one with the highest inhibitory efficiency was selected for subsequent experiments. The shRNA- DNTTIP2 plasmid (sh-DNTTIP2), a control non-targeting plasmid (NC), and the overexpressing plasmid (oe-DNTTIP2) were transfected into 70%-confluent cells using Lipofectamine 3000, in accordance with the supplied protocol. The overexpression, knockdown, and transfection efficiency were evaluated using Western blotting and GFP expression.
Colony-Formation Assay
Colony-forming assays were conducted according to published protocols.11
Wound Healing Assay
The method used was based on a published protocol.12 The GFP levels in the cells on the membrane lower surface were expressed as means ± SEM from three separate experiments.
Transwell Assay
The capability of cell invasion was examined with a Transwell assay as previously described.13
Co-Culture System
THP-1 monocytic cells (1 × 105 cells/mL) were incubated with 10 ng/mL Phorbol-12-myristate-13-acetate (PMA) for 48 h to induce their differentiation into M0 macrophages. Then, the medium was removed and replaced with serum free DMEM and then the cells were allowed to grow for 24 h. To mimic the interactions between macrophages and tumors, 5×105 glioma cells were placed in the upper chamber of a 6-well 0.4um pore size Transwell apparatus and the M0 macrophages were placed in the lower chamber. The macrophages in the lower chamber were collected after 48 h and stained by immunofluorescence.
Immunofluorescence Staining
Immunofluorescence staining followed the method described in our previous study.14
Endothelial Cell Tube Formation Assay
Tube formation assays are useful for assessing the angiogenic capacity of cells.15 Here, this was done using the branch point method. In brief, glioma cells were grown in the upper chambers of 24-well Transwell apparatuses 0.4 µm pore size) and HUVEC cells (5×104/well) were grown in the lower chambers. The plates were pre-coated with Matrigel (50 µL) and cultured at 37°C in a 5% CO2 incubator. After 12 h, the HUVECs were incubated with 1 mM Calcein-acetoxymethyl ester (calcein-AM) for 1 h at 37°C and examined under fluorescence microscopy (Olympus CKX-41, Japan).
Xenograft Tumor Model
The animal experimental protocol (ethics number: 2021-10-029) was approved by the ethics committee of the Jiangsu Province Hospital of Chinese Medicine. U251 cells transfected with sh-DNTTIP2, oe-DNTTIP2, as well as NC cells (1 × 107 cell/mouse) were injected subcutaneously into the right armpit region (n = 6 per group). The presence of tumors was visible after seven days. Tumor diameters (maximum and minimum) were measured twice a week. The mice were euthanized after 35 days and the sera and tumors were excised. Tumor volumes were calculated as V = 1/2ab2, and tumor growth curves were drawn.
Statistical Analysis
Data are expressed as means ± (SEM). All experiments were carried out thrice, independently. ****P < 0.0001, ***P < 0.001, **P < 0.01 and *P < 0.05 were defined to be statistically significant.
Data Acquisition
Datasets of LGG patients, including both genetic and clinical information, were obtained from The Cancer Genome Atlas (TCGA) and the Chinese Glioma Genome Atlas (CGGA).16 In total, information on 530 tumor and 0 normal tissues were downloaded from the TCGA, and 182 tumor and 0 normal tissues from the CGGA. To study the functions of DNTTIP2 and its immune relationships, two groups were established based on DNTTIP2 expression according to TCGA-LGG. In addition, the datasets GSE70630, GSE84465, GSE135437 and GSE148842 were used to conduct analysis at the single-cell level.17
Data Analysis Tools
Differences in DNTTIP2 levels were analyzed using ANOVA and the t-test. Changes in DNTTIP2 frequency in the TCGA data were analyzed using the CBioportal database.18 The R packages “survival,” and “survminer” were used for Kaplan–Meier survival analysis.19 For functional enrichment analysis and Gene Set Variation Analysis (GSVA), the R package “Limma” was used to identify differentially expressed genes and differentially enriched genesets between DNTTIP2 and Metascape as well as “GSVA” package were used to conduct enrichment analysis.20,21
Investigation of Immune Cell Infiltration
CIBERSORT high-performance computation for measuring the cellular constituents in overall tissue datasets22 was used to estimate immune infiltration. All results were implemented by the “ggplot2” and “pheatmap” packages in R.23 We investigated divergences in immune cell populations between the two DNTTIP2 high and low expression groups. The GSVA algorithm was used to calculate the correlation coefficient between DNTTIP2 and the abundance of immune cell infiltration.24 Subsequently, GSE28238 and GSE45921 were further conducted to evaluate the relationship between DNTTIP2 levels and M2 macrophage abundance based on the CIBERSORT algorithm.25
Results
Prognosis Analysis of DNTTIP1/2 mRNA Expression in Pan-Cancers
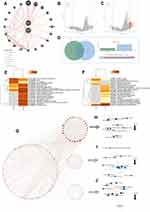
We performed a pan-cancer survival analysis based on the GEPIA database to evaluate the DNTTIP1/2 prognostic values. In the above analyses, we found that DNTTIP1/2 mRNA expression was associated clinical outcome (OS: Overall survival; DFS: Disease-free survival) of patients with Liver hepatocellular carcinoma (LIHC) and LGG (Figure 2A and B).
A Kaplan-Meier survival analysis based on TCGA demonstrated that high DNTTIP1/2 levels correlated with significantly worse patient survival rates in LGG (Figure 2C) and LIHC (Figure S1A). The expression profiles and clinical characteristics of LGG and LIHC patients were obtained from TCGA to probe the relationship between DNTTIP1/2 expression status and clinicopathological characteristics (Figure 2D for LGG and Figure S1B for LIHC). In order to evaluate the prognostic value, the clinical and pathological features, including WHO grade, Isocitrate dehydrogenase (IDH) status, 1p19q codeletion, primary treatment outcome for LGG and LHIC ‘tumor-node-metastasis” (TNM) stage, pathological stage, serum AFP, histological grade and TNM stages, were synthesized. Subsequently, the prognostic value of DNTTIP1/2 expression status in LGG was validated in CGGA (Figures S2 and S3). Results from the TGGA and CGGA databases demonstrated that DNTTIP1/2 expression status in LGG can have prognostic value, and have been associated with the genomic features of LGG. This part of the work is the process of screening the cancer types under study.
Immunohistochemical Staining of DNTTIP2
DNTTIP2 protein expression in LGG and normal brain tissue was further verified using immunohistochemistry (IHC) data acquired from the HPA database (http://www.proteinatlas.org/), which revealed that although DNTTIP2 protein was not expressed or low expressed in normal brain tissue, low expression was evident in LGG tissues (Figure S4).
Functional Enrichment Analysis of DNTTIP1/2
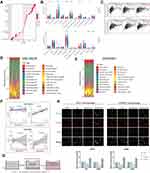
Figure 3A showed the interactions of DNTTIP1/2 predicted by GeneMANIA. These predicted interactions include genetic and physical interactions, co-localization and co-expression, as well as the likelihood of common protein domains and pathways. We obtained genes co-expressed with DNTTIP1/2 in TCGA-LGG based on the “Limma” R package (Figure 3B and C). Then, we identified 38 common genes co-expressed with DNTTIP1 and DNTTIP2 based on intersection analysis (Figure 3D). Furthermore, using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis, the top 20 GO and KEGG categories ranked according to P-value are shown in Figure 3E and F, suggesting that DNTTIP2 may be involved in more cancer-related pathways compared to DNTTIP1. We noted that the enriched GO and KEGG terms of DNTTIP2 associated with “immune effector process”, “blood vessel development”, “extracellular matrix”, “regulation of cytokine production” and “Complement and coagulation cascades”.
After removing the co-expressed genes that were not assigned, we created a Protein-Protein Interaction network (PPI) based on the STRING database (Figure 3G) and then the key subnetwork obtained by the molecular complex detection (MCODE) plug-in showed even stronger association with macrophage activation and angiogenesis (Figure 3H–J).
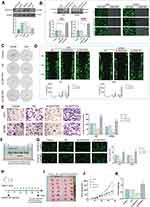
DNTTIP2 Promotes Malignant Phenotypes in Glioma
The expression of DNTTIP2 was further explored in the glioma cells by Western blot (Figure 4A). The DNTTIP2 protein level was the highest in U87MG and U251, therefore these two cell lines were used for further experiments, with GFP expression and Western blot verifying transfection efficiency (Figure 4B). DNTTIP2 silencing decreased the capacity of forming the tumor cell clone formation, migration, invasion and tube formation in vascular endothelial cells (Figure 4C–G). Moreover, the stable DNTTIP2 overexpression in the U251 cells promoted the formation of the subcutaneous xenograft tumors in vivo (Figure 4H–K).
Immune Microenvironment Analysis
In this section, we explored the role of DNTTIP2 in the LGG immune cell infiltration. First, the ssGSEA algorithm being used to access the correlation between DNTTIP2 levels and the degree of infiltration degree several immune cell types showed that DNTTIP2 levels were positively associated with the abundance of macrophages (Figure 5A). Then, twenty-two profiles of immune cells based on “CIBERSORT” algorithm were constructed to detect the relationship between the DNTTIP2 level and immune cells levels (Figure 5B). The DNTTIP2 level was found to correlate with B cell native, B cell plasma, T cell CD4+ native, T cell CD4+ memory resting, T cell follicular helper, macrophage M2, eosinophil, and neutrophil cells. In addition, The TIMER web tool was used to explore potential correlations between DNTTIP2 levels and immune cell infiltration, revealing that the DNTTIP2 levels were correlated with CD4+T cells (R = 0.373, P = 3.69e-17), macrophages (R = 0.428, P = 1.63e-22), Neutrophil (R = 0.552, P = 2.66e-39), dendritic cells (R = 0.512, P = 3.67e-33), and B cells (R = 0.55, P = 4.63e-39) (Figure 5C). These findings indicate close relationships between DNTTIP2 and immune cell infiltration in LGG.
Then we assessed the relationship between DNTTIP2 and macrophage polarization based on the GSE28238 and GSE45921 datasets. The 22 immune cell profiles for LGG cases were presented in Figure 5D and E after filtering out normal samples. Further calculations showed that in GSE28238 and GSE45921, DNTTIP2 expression levels were positively associated with M2 macrophage abundance (Figure 5F). To further detect the effect of DNTTIP2 expression on the abundance of M2 macrophages, we established a tumor-macrophage co-culture model as shown in the Figure 5G and observed that DNTTIP2 knockdown significantly downregulated the levels of CD206 and CD163 which are now considered to be indicated markers for M2 tumor-associated macrophages (TAMs) (Figure 5H).
Taken together, these findings indicate that DNTTIP2 levels are correlated with the infiltration of immune cells, and are especially associated with the infiltration of M2 macrophages.
GSVA and Single-Cell Level Analysis of DNTTIP2
To further detect the potential function of DNTTIP2 in LGG, we conducted a GSVA. The distribution of GSVA scores for the LGG samples in TCGA and CGGA is shown in Figure S4A and B. The results of the differential score analysis based on the median DNTTIP2 levels showed that “TNFA SIGNALING VIA NFKB” and “IL6 JAK STAT3 SIGNALING” were activated in the high-DNTTIP2, in both cohorts (Figure 6A and B).
We also performed single-cell expression analysis to explore DNTTIP2 functions in LGG. With the GSVA, we concentrated on “TNFA SIGNALING VIA NFKB” and “IL6 JAK STAT3 SIGNALING”. As shown in Figure 6C–F, the Gene Set Enrichment Analysis (GSEA) results suggested that cells expressing DNTTIP2 tend to express genes related to “ANGIOGENESIS” and “IL-6 JAK STAT3 SIGNALING”. This may be the mechanism of DNTTIP2 in promoting LGG development.
Methylation and Mutation Analysis of DNTTIP2
Considering the importance of methylation as an epigenetic modification, we next investigated whether DNTTIP2 expression levels were affected by DNTTIP2 DNA methylation in LGG. In the TCGA data, we found that the methylation results from 12 probes, cg08526814, cg19164253, cg23110891, cg23633626, cg19906608, cg23015158, cg23956358, cg01139627, cg15589930, cg16398686, cg16833230, and cg26688893, were associated with DNTTIP2 expression (Figure S6B). The survival analysis of all the methylation probes of DNTTIP2 is shown in Figure S6F.
The mutation frequencies in three groups of LGG patients were analyzed through the cBioPortal database (Figure S6C). DNTTIP2 showed a low rate of mutation (∼0.2%) and a low amplification rate of ∼0.4% of DNTTIP2 in the TCGA LGG cases. There was a statistically significant link between DNTTIP2 Copy number variations (CNV) and overall survival in LGG patients (Figure S6D) as well as gene expression (Figure S6E). The DNTTIP2 CNV and immune cell infiltration were also associated with each other (Figure S6F).
Discussion
Due to the highly infiltrative growth pattern of gliomas, the heterogeneity within the tumor, the poor demarcation from normal brain tissue, and the lack of effective indicators for clinical monitoring, the patient has both a significant risk of tumor recurrence and a very poor prognosis after surgery.26 In addition, the treatment of LGG is more complex and difficult than that of extracerebral solid tumors, and many conventional chemotherapeutic agents have difficulty crossing the intact blood-brain barrier, thus affecting the application and efficacy of chemotherapeutic agents.27 Therefore, exploring specific biomarkers associated with glioma tumor diagnosis, grading and staging, to guide the use of drugs and prognosis is of great clinical importance to improve the prognosis of glioma. The Kaplan-Meier survival analysis demonstrated that high levels of DNTTIP1/2 were linked to poor prognosis in LGG patients. Interestingly, we found that DNTTIP1/2 expression levels were linked to multiple clinical features such as tumor grade, IDH mutation, 1p/19q codeletion, suggesting the clinical relevance of DNTTIP1/2.
DNTTIP is also known as Terminal Deoxynucleotidyltransferase (TDT) -Interacting factor, elevated activity of TDT was found in more than 90% of leukemic cells in acute lymphoblastic leukemia as well as approximately 30% of leukemic cells in chronic granulocytic Leukemia crisis.28,29 TDT levels are now thought to be associated with poor prognosis, chemotherapy response and shortened survival.30 Considering the significant pro-cancer effect of TDT, we speculate that DNTTIP may play a similar role in tumors. We first performed functional enrichment analysis, which showed that, compared to DNTTIP1, DNTTIP2 may involve a wider range of biological processes, including immune effector process, extracellular matrix, and blood vessel development. Notably, the distinctive feature of glioma growth and progression is the development of a large number of blood vessels that are highly aberrant and dysfunctional, with detrimental consequences for the therapeutic outcome,31,32 while the extracellular matrix is enriched with molecules that have the ability to provide support structures for the embedded vessels and to signal surrounding cells to initiate or terminate the angiogenic process.33,34 Subsequent vitro and vivo experiments verified that DNTTIP2 significantly promoted the proliferation and activation of glioma cells and tumor growth, and glioma cells with high expression of DNTTIP2 have a strong ability to promote angiogenesis.
It has been reported that in the late stages of glioma growth, host cells located in the vicinity of blood vessels are recruited in large numbers, which are identified by selective immune markers as predominantly macrophages with a very small proportion of T cells, B cells, natural killer cells, neutrophils and dendritic cells.35,36 The vascular phenotype, characterized by blood vessel diameter increase, leakiness and loss of branching complexity, is mainly accompanied by the secretion of large amounts of VEGFA by M2-like macrophages in the late stages of glioma growth and relocalization in the vicinity of vascular structures.37,38 Furthermore, it has been demonstrated that the number of M2 macrophages is significantly and positively correlated with vessel diameter in accordance with the WHO classification, and that macrophage depletion treatment can drive improvements in vessel structure and perfusion.39,40 These works are revealing the fundamental fact that glioma vascular network formation and macrophage recruitment are highly consistent. Starting from this fact, while the enrichment analysis also suggested a significant correlation between DNTTIP2 and the immune effector process, we found a significant positive correlation between DNTTIP2 levels and the abundance of M2 macrophage infiltration calculated by the GSVA and cibersort algorithms based on three independent data sets. Subsequently, vitro experiments based on co-culture system directly confirmed the powerful ability of DNTTIP2 to promote the proliferation of M2 macrophages.
Several underlying mechanisms might be involved in the effect of DNTTIP2 in LGG. We preliminarily explored the molecular mechanisms of DNTTIP2 based on GSVA. The results demonstrated that “TNFA SIGNALING VIA NF-KB” and “IL-6 JAK STAT3 SIGNALING” were significantly activated in the DNTTIP2 high-expression group. Subsequent extensive single-cell level analysis also confirmed that a proportion of glioma cells expressing DNTTIP2 were also more inclined to express angiogenesis-related genes and “IL-6 JAK STAT3 SIGNALING”. However, a complex disease such as glioma generally does not result from mutations or dysfunctions of a single gene, but from abnormalities in the function of the associated regulatory network. The activation of a single gene or pathway may not explain the full extent of its disease progression, and this part of our work only provides a potential direction for subsequent researchers, the mechanisms involved of which will require extensive experimental validation.
Alterations in DNA are inextricably linked with carcinogenesis.41 Although we did not identify an association between DNTTIP2 mutation and LGG prognosis, we found that DNTTIP2 CNVs were correlated with DNTTIP2 levels and LGG patients’ prognosis. In addition, hypomethylation of several DNTTIP2 probes (cg23110891, cg19164253, cg23956358, cg01139627, cg16833230, and cg15589930) and hypermethylation of one probe (cg23015158) were associated with raised DNTTIP2 expression and reduced patient survival. Considering that different region of methylation sites has very different effects on gene expression, future studies should focus on the effects of specific DNTTIP2 methylation sites on both expression of the gene and patient mortality.
Conclusion
In summary, this study explored the correlation between DNTTIP1/2 and the prognostic and clinical characteristics of LGG based on TCGA and CGGA and showed that raised DNTTIP1/2 expression was linked to unfavorable prognosis. From this point, the potential biological functions of DNTTIP2 were explored by functional enrichment and immune infiltration analysis, and the results showed that DNTTIP2 could significantly affect the degree of activation of LGG M2 macrophages as well as promote angiogenesis, and this effect might be associated with IL6/JAK/STAT3 signaling. Finally, we also found that CNVs and DNA methylation may be responsible for the overexpression of DNTTIP2 in LGG. Few studies have shown an association between DNTTIP2 and LGG or other cancers, and the present study may provide a basis for future development of therapeutic strategies for LGG. The main limitation of our study is that we do not have samples of glioma patients for clinical characterization and validation, and our team is still conducting follow-up studies to elucidate the mechanism.
Data Sharing Statement
All data relevant to this study were obtained from publicly reported, open, and online data sources. We declare that all the data in this article are authentic, valid, and available upon reasonable request.
Ethical Statement
This study was considered exempt due to the use of publicly available, de-identified data. The study was granted a waiver from approval by the Ethical Committee of the Jiangsu Province Hospital of Chinese Medicine.
Acknowledgments
Thanks to each author for their important contribution and thanks to all the participants. We sincerely acknowledge the public databases: GEO, CGGA, TCGA, and GEPIA. Yuan-jie Liu and Shu-hong Zeng are co-first authors. Jie-pin Li and Ying-ying Zhu are designated conjointly as corresponding authors.
Author Contributions
All authors made a significant contribution to the work reported, whether in study conception, design, and execution; data acquisition, analysis, and interpretation; or all these areas. All authors were involved in drafting, revising, or critically reviewing the article; gave final approval for the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work. Yuan-jie Liu and Shu-hong Zeng are co-first authors. Dr. Ying-ying Zhu and Dr. Jie-pin Li are designated conjointly as corresponding authors.
Funding
The present study was supported by the Youth Science and Technology Project of Suzhou (No. KJXW2019059); the Suzhou Science and Technology Development Plan (No. SYSD2019006) and (No. SYSD2019008); Clinical Medicine Science and Technology Development Fund of Jiangsu University (No. JLY20180146).
Disclosure
The authors have declared no conflicts of interest in this work.
References
1. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. doi:10.1007/s00401-016-1545-1
2. Wesseling P, Capper D. WHO 2016 classification of gliomas. Neuropathol Appl Neurobiol. 2018;44(2):139–150. doi:10.1111/nan.12432
3. Adilijiang A, Hirano M, Okuno Y, et al. Next generation sequencing-based transcriptome predicts bevacizumab efficacy in combination with temozolomide in glioblastoma. Molecules. 2019;24(17):3046. doi:10.3390/molecules24173046
4. Itoh T, Fairall L, Muskett FW, et al. Structural and functional characterization of a cell cycle associated HDAC1/2 complex reveals the structural basis for complex assembly and nucleosome targeting. Nucleic Acids Res. 2015;43(4):2033–2044. doi:10.1093/nar/gkv068
5. Yamashita N, Shimazaki N, Ibe S, et al. Terminal deoxynucleotidyltransferase directly interacts with a novel nuclear protein that is homologous to p65. Genes Cells. 2001;6(7):641–652. doi:10.1046/j.1365-2443.2001.00449.x
6. Farooqi AA, Fuentes-Mattei E, Fayyaz S, et al. Interplay between epigenetic abnormalities and deregulated expression of microRNAs in cancer. Semin Cancer Biol. 2019;58:47–55. doi:10.1016/j.semcancer.2019.02.003
7. Mrakovcic M, Bohner L, Hanisch M, et al. Epigenetic targeting of autophagy via HDAC inhibition in tumor cells: role of p53. Int J Mol Sci. 2018;19(12):3952. doi:10.3390/ijms19123952
8. Bantscheff M, Hopf C, Savitski MM, et al. Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nat Biotechnol. 2011;29(3):255–265. doi:10.1038/nbt.1759
9. Sawai Y, Kasamatsu A, Nakashima D, et al. Critical role of deoxynucleotidyl transferase terminal interacting protein 1 in oral cancer. Lab Invest. 2018;98(8):980–988. doi:10.1038/s41374-018-0070-3
10. Kuriakose S, Onyilagha C, Singh R, et al. TLR-2 and MyD88-dependent activation of MAPK and STAT proteins regulates proinflammatory cytokine response and immunity to experimental trypanosoma congolense infection. Front Immunol. 2019;10:2673. doi:10.3389/fimmu.2019.02673
11. Yu S, Hu Q, Fan K, et al. CSNK2B contributes to colorectal cancer cell proliferation by activating the mTOR signaling. J Cell Commun Signal. 2021;15(3):383–392. doi:10.1007/s12079-021-00619-1
12. Han JH, Yoon JS, Chang D-Y, et al. CXCR4-STAT3 axis plays a role in tumor cell infiltration in an orthotopic mouse glioblastoma model. Mol Cells. 2020;43(6):539–550. doi:10.14348/molcells.2020.0098
13. Cao R, Wang G, Qian K, et al. Silencing of HJURP induces dysregulation of cell cycle and ROS metabolism in bladder cancer cells via PPARγ-SIRT1 feedback loop. J Cancer. 2017;8(12):2282–2295. doi:10.7150/jca.19967
14. Liu Y-J, Zeng S-H, Hu Y-D, et al. Overexpression of NREP promotes migration and invasion in gastric cancer through facilitating epithelial-mesenchymal transition. Front Cell Dev Biol. 2021;9:746194. doi:10.3389/fcell.2021.746194
15. Yang Z, Li H, Luo P, et al. UNC5B promotes vascular endothelial cell senescence via the ROS-mediated P53 pathway. Oxid Med Cell Longev. 2021;2021:5546711. doi:10.1155/2021/5546711
16. Serrano-Garrido O, Peris-Torres C, Redondo-García S, et al. ADAMTS1 supports endothelial plasticity of glioblastoma cells with relevance for glioma progression. Biomolecules. 2020;11(1):44. doi:10.3390/biom11010044
17. Tan YQ, Li YT, Yan TF, et al. Six immune associated genes construct prognostic model evaluate low-grade glioma. Front Immunol. 2020;11:606164. doi:10.3389/fimmu.2020.606164
18. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi:10.1126/scisignal.2004088
19. Ranstam J, Cook JA, Ranstam J, Cook JA. Kaplan-Meier curve. Br J Surg. 2017;104(4):442. doi:10.1002/bjs.10238
20. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi:10.1093/nar/gkv007
21. Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi:10.1038/s41467-019-09234-6
22. Kawada J-I, Takeuchi S, Imai H, et al. Immune cell infiltration landscapes in pediatric acute myocarditis analyzed by CIBERSORT. J Cardiol. 2021;77(2):174–178. doi:10.1016/j.jjcc.2020.08.004
23. Fang Y, Huang S, Han L, et al. Comprehensive analysis of peritoneal metastasis sequencing data to identify LINC00924 as a prognostic biomarker in gastric cancer. Cancer Manag Res. 2021;13:5599–5611. doi:10.2147/CMAR.S318704
24. Hu B, Yang XB, Sang XT. Development and verification of the hypoxia-related and immune-associated prognosis signature for hepatocellular carcinoma. J Hepatocell Carcinoma. 2020;7:315–330. doi:10.2147/JHC.S272109
25. Zeng WJ, Cheng Q, Wen Z-P, et al. Aberrant ASPM expression mediated by transcriptional regulation of FoxM1 promotes the progression of gliomas. J Cell Mol Med. 2020;24(17):9613–9626. doi:10.1111/jcmm.15435
26. Sturm D, Pfister SM, Jones DTW. Pediatric gliomas: current concepts on diagnosis, biology, and clinical management. J Clin Oncol. 2017;35(21):2370–2377. doi:10.1200/JCO.2017.73.0242
27. Tang W, Fan W, Lau J, et al. Emerging blood-brain-barrier-crossing nanotechnology for brain cancer theranostics. Chem Soc Rev. 2019;48(11):2967–3014. doi:10.1039/c8cs00805a
28. Drexler HG, Sperling C, Ludwig WD. Terminal deoxynucleotidyl transferase (TdT) expression in acute myeloid leukemia. Leukemia. 1993;7(8):1142–1150.
29. Patel KP, Khokhar FA, Muzzafar T, et al. TdT expression in acute myeloid leukemia with minimal differentiation is associated with distinctive clinicopathological features and better overall survival following stem cell transplantation. Mod Pathol. 2013;26(2):195–203. doi:10.1038/modpathol.2012.142
30. Di Santo R, Maga G. Human terminal deoxynucleotidyl transferases as novel targets for anticancer chemotherapy. Curr Med Chem. 2006;13(20):2353–2368. doi:10.2174/092986706777935087
31. Shi Z, Chen Q, Li C, et al. MiR-124 governs glioma growth and angiogenesis and enhances chemosensitivity by targeting R-Ras and N-Ras. Neuro Oncol. 2014;16(10):1341–1353. doi:10.1093/neuonc/nou084
32. Plate KH, Scholz A, Dumont DJ. Tumor angiogenesis and anti-angiogenic therapy in malignant gliomas revisited. Acta Neuropathol. 2012;124(6):763–775. doi:10.1007/s00401-012-1066-5
33. Marchand M, Monnot C, Muller L, et al. Extracellular matrix scaffolding in angiogenesis and capillary homeostasis. Semin Cell Dev Biol. 2019;89:147–156. doi:10.1016/j.semcdb.2018.08.007
34. Ferrer VP, Moura Neto V, Mentlein R. Glioma infiltration and extracellular matrix: key players and modulators. Glia. 2018;66(8):1542–1565. doi:10.1002/glia.23309
35. Hambardzumyan D, Gutmann DH, Kettenmann H. The role of microglia and macrophages in glioma maintenance and progression. Nat Neurosci. 2016;19(1):20–27. doi:10.1038/nn.4185
36. Wei J, Chen P, Gupta P, et al. Immune biology of glioma-associated macrophages and microglia: functional and therapeutic implications. Neuro Oncol. 2020;22(2):180–194. doi:10.1093/neuonc/noz212
37. Wick W, Platten M, Wick A, et al. Current status and future directions of anti-angiogenic therapy for gliomas. Neuro Oncol. 2016;18(3):315–328. doi:10.1093/neuonc/nov180
38. Segura-Collar B, Garranzo-Asensio M, Herranz B, et al. Tumor-derived pericytes driven by EGFR mutations govern the vascular and immune microenvironment of gliomas. Cancer Res. 2021;81(8):2142–2156. doi:10.1158/0008-5472.CAN-20-3558
39. Zhu C, Kros JM, Cheng C, et al. The contribution of tumor-associated macrophages in glioma neo-angiogenesis and implications for anti-angiogenic strategies. Neuro Oncol. 2017;19(11):1435–1446. doi:10.1093/neuonc/nox081
40. Daubon T, Hemadou A, Romero Garmendia I, et al. Glioblastoma immune landscape and the potential of new immunotherapies. Front Immunol. 2020;11:585616. doi:10.3389/fimmu.2020.585616
41. Skvortsova K, Stirzaker C, Taberlay P, Blewitt M. The DNA methylation landscape in cancer. Essays Biochem. 2019;63(6):797–811. doi:10.1042/EBC20190037
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.