Back to Journals » OncoTargets and Therapy » Volume 13
DNMT1 Enhances the Radiosensitivity of HPV-Positive Head and Neck Squamous Cell Carcinomas via Downregulating SMG1
Authors Zhang C, Mi J, Deng Y, Deng Z, Long D, Liu Z
Received 15 August 2019
Accepted for publication 18 February 2020
Published 15 May 2020 Volume 2020:13 Pages 4201—4211
DOI https://doi.org/10.2147/OTT.S227395
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Chunlin Zhang,1,* Jiaoping Mi,2,* Yuan Deng,1 Zeyi Deng,3 Dan Long,1,4 Zhaohui Liu1
1Department of Otorhinolaryngology, Head and Neck Surgery, Affiliated Hospital of Zunyi Medical University, Zunyi, People’s Republic of China; 2Department of Otorhinolaryngology, Head and Neck Surgery, The Fifth Affiliated Hospital of Sun Yat-Sun University, Zunyi, People’s Republic of China; 3Department of Otorhinolaryngology, Head and Neck Surgery, Zhujiang Hospital, Southern Medical University, Guangzhou, People’s Republic of China; 4The Graduate School of Zunyi Medical University, Zunyi, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Chunlin Zhang; Zhaohui Liu
Department of Otorhinolaryngology, Head and Neck Surgery, Affiliated Hospital of Zunyi Medical University, Zunyi, People’s Republic of China
Email [email protected]; [email protected]
Introduction: Head and neck squamous cell carcinoma (HNSCC), which rank the 7th malignant tumors worldwide, is closely related to methylation and HPV infection. Ionizing radiation therapy is the main strategy for HNSCC patients in advanced stage. Previously, HPV-positive HNSCC predict better prognosis than HPV-negative HNSCCs under radiotherapy, however its molecular mechanism is unresolved. SMG1 serves as a potential tumor suppressor in various cancers, including HNSCC.
Methods: The mRNAs and proteins expression of HPV E6/E7, p16, p53, DNMT1, SMG1 were detected after different treatments by qPCR and Western blot. The clone formation ability was measured in radiation dose after different treatments.
Results: In our study, the expression of HPV16 E6, DNA Methyltransferase 1(DNMT1) and SMG1 in head and neck carcinomas cell lines was detected by RT-qPCR and Western blot. Forced E6 level in HPV-negative cells by overexpression plasmid promoted the expression of DNMT1, which resulted in decreased SMG1 expression. Silenced SMG1 in HPV-negative HNSCC cells elicited increased radiation sensitivity, suggesting that SMG1 may be an effective switch to regulate the effect of radiotherapy in HNSCC.
Conclusion: Our study indicated that DNMT1 enhances the radiosensitivity of HPV-positive head and neck squamous cell carcinomas via downregulating SMG1.
Keywords: head and neck cancer, HPV, DNMT1, SMG1, radiotherapy
Introduction
Head and neck cancer is one of the most common seem malignant tumors worldwide, about 600,000 new cases of patients are diagnosed every year.1 This kind of tumor originates in nasopharynx, sinonasal tract, larynx, hypopharynx, oropharynx and mucosa lining the oral cavity.2 About 10% of all patients arise in the oropharynx. The most common kind of head and neck cancer is squamous cell carcinoma (HNSCC).3 HNSCC is concealed and more than 60% of patients are in advanced stage at the time of first visit.4 Risk factors for head and neck cancer are complex including genetic background, smoking, drinking and biological factors such as virus, physical and chemical factors, etc.5 Although the current multidisciplinary treatments based on surgery, radiotherapy and chemotherapy, and targeted therapy have made great progress, the overall survival rate of patients has not been shown improved in recent decades. The 5-year survival rate is only 40%-50%.
Recent studies have shown that the occurrence of multiple tumors in humans is associated with human papillomavirus (HPV) infection.6–8 HPV carcinogenicity has been shown to play an important role in the etiology of genitourinary tumors in women.9 It is currently believed that HPV infection is also a relevant causative factor in the development of SCC of the head and neck.10 Some surveys have shown that 20%-25% of HNSCC patients are positive for HPV, especially oropharyngeal cancer.11 HPV causes tumorigenesis by expressing E6 and E7 proteins.12 E6 and E7 can degrade the expression products of the tumor suppressor genes p53 and pRb.13 HPV E6 interacts with P53, which degrades P53 protein and loses its anti-cancer effect, increasing the chance of host cell malignant transformation.13 HPV E7 acts on Rb to inactivate it.14 The inactivated Rb gene can reversely activate multiple transcription factors. The binding of DNA cis-acting elements activates transcription of p16 gene, resulting in high activation of p16 protein.15 The expression level of p16 and Ki-67 is increased, which causes cell cycle disorder and causes cell malignant transformation. Recent studies have shown that E6 and E7 can also bind to other proteins, such as Bak and p21, leading to their genetic instability.16 However, the separate expression of E6 and E7 is not sufficient to cause malignant transformation of cells, and the mechanism of genetic alteration caused by virus-induced genomic instability remains unclear. In addition, HPV-positive patients and HPV-negative patients have different responses to treatment and prognosis.17 HPV-positive patients have more obvious effects on radiotherapy.
DNA methyltransferase 1 (DNMT1) encodes an enzyme that transfers methyl group to cytosine nucleotides of genomic DNA. This protein is the major enzyme responsible for maintaining methylation patterns following DNA replication and shows a preference for hemimethylated DNA. Methylation of DNA is an important complement of mammalian epigenetic gene regulation.18 Aberrant methylation patterns are found in human tumors and associated with developmental abnormalities.19 SMG1 is a protein involved in nonsense-mediated mRNA decay (NMD) as part of the mRNA surveillance complex. The protein has kinase activity and is thought to function in NMD by phosphorylating the regulator of nonsense transcripts protein.20 It has been reported that DNMT1 affects the expression of SMG1 and changes the sensitivity to radiotherapy in tumors. But it has not been widely studied in head and neck neoplasm.
In this study, we hypothesized that HPV E6 would down-regulate SMG1 by increasing the expression of DNMT1. The present research aimed to find the underlying mechanisms that affected the sensitivity of head and neck tumors to radiation therapy, in order to improve the effect of cancer patients on radiation therapy and improve survival rate.
Materials and Methods
Cell Cultures and Transfection
We used 2 HPV-negative (FaDu, UM-SCC-4) and 2 HPV-positive (UPCI-SCC-090, UM-SCC-47) HNSCC cell lines which were bought from ATCC immortalized by expression of hTERT. FaDu and UM-SCC-47 cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum. UM-SCC-4 cell line was cultured in DMEM/F12 medium (Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum and 0.4% hydrocortisone. UPCI-SCC-090 cell line was cultured in minimum Eagle’s medium (Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum. All the cells were cultured at 37°C in a humidified atmosphere of 5% CO2. SiRNAs were purchased from RiboBio (Guangzhou, Chian) and plasmids were purchased from GeneChem. SiRNAs and plasmids transfection were performed using Lipofectamine 3000 transfection reagent (Invitrogen) following the manufacturer’s protocol. After 48 h of transfection, the cells were collected and used for further analysis.
Clonogenic survival assay: Cells were seeded in 6-well plates at different densities depending on the dose of radiotherapy and treated with a dose of 2, 4, 6, 8 Gy over an appropriate field size. After 14 days culture, the colonies were fixed and stained with methylene blue in methanol. The colonies containing at least 50 cells were counted and survival fractions were corrected for the plating efficiencies.
Quantitative Real-Time PCR
Total RNA was extracted from cells using Trizol reagent (Invitrogen, Carlsbad, CA), and the reverse-transcription reactions were performed using random primers and an M-MLV Reverse Transcriptase kit (Invitrogen, Carlsbad, CA). Real-time PCR was performed to use a standard SYBR Green qPCR master mix kit (DBI, German) protocol on Line Gene 9600 Plus Real Time PCR system (Bioer, China) according to the instructions. GAPDH was used as reference. The 2−ΔΔCt method was used to determine the relative quantitation of gene expression levels. Each sample was analyzed in triplicate. Primer sequences were listed in Table 1.
 |
Table 1 The Primer Sequences for qPCR |
Western Blot
Total protein was extracted from the cells by lysis with RIPA Lysis Buffer (DGCS Biotechnology, China). The protein content of the lysate was then determined using the BCA kit (Beyotime, China) according to the manufacturer’s protocol. Cell lysates were resolved on a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto a polyvinylidene fluoride (PVDF) membrane (Millipore, USA). The membrane was blocked with TBS-T (0.1% Tween-20 in TBS) containing 5% BSA and incubated with primary antibody overnight at 4°C. After three washes with TBS-T, the membrane was incubated with HRP-conjugated secondary antibody for 1 h and then washed with TBS-T. The immune complexes were detected by chemiluminescence (Supersignal West Femto; Pierce Biotechnology) using MyECL Imager (Thermo Scientific). Mean densitometry data from independent experiments were normalized to the control. Primary antibodies are listed as follows: E6 (ab70, abcam, USA), E7 (sc-51951, santa cruz, USA), p16 (sc-377412, santa cruz, USA), p53 (sc-71819, santa cruz, USA), DNMT1 (5032s, CST, USA), SMG1 (4993s, CST, USA), GAPDH (60004-1-Ig, proteintech, USA). All experiments were repeated three times.
Statistical Analysis
All data analysis was performed using SPSS 23.0 software. Given that the study data were consistent with a normal distribution and the variance was equal, the differences between two groups were calculated using Student’s t-tests, and the differences between more than two groups were analyzed using One-way ANOVA; adjusted P-values were calculated by Tukey comparison test. All data were presented as the mean ± SD. A P-value<0.05 was considered statistically significant.
Result
HPV-Positive HNSCC Cell Lines are More Sensitive to Radiation Than HPV-Negative HNSCC Cell Lines
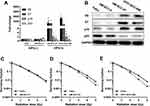
According to previous studies, FaDu and UM-SCC-4 are classic HPV-negative HNSCC cell lines that do not express HPV16 E6, while UM-SCC-47 and UPCI-SCC-090 are deemed as HPV-positive cell lines that express HPV16 E6. E6 and E7 were significantly higher in UM-SCC-47 and UPCI-SCC-090 than FaDu and UM-SCC-4 (Figure 1A) by RT-qPCR. In addition, the trends of p16 were the same as that of E6 and p53 was down-regulated on HPV-positive cells than HPV-negative cells (Figure 1A). The expression of the corresponding proteins was consistent with the mRNA (Figure 1B). With regard to radiosensitization, HNSCC cells were treated with different doses of radiation, and the survival rates were assessed by colony formation assay. In parallel with previous studies, UM-SCC-47 and UPCI-SCC-090 showed more radiation sensitivity than those of FaDu and UM-SCC-4 (Figure 1C–E). From the above results, we concluded that HPV-positive cells were more sensitive to radiation than HPV-negative cells.
HPV-16 E6 Promotes the Sensitivity of Radiation in HNSCC Cells
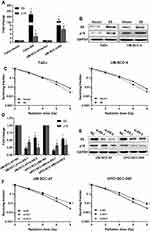
To investigate whether the HPV E6 level contributes to increased sensitivity of HNSCC to radiation, we constructed E6 overexpression FaDu and UM-SCC-4 cells and E6 silenced UM-SCC-47 and UPCI-SCC-090 cells and assessed sensitivity of the cells to radiation in clonogenic survival assay (Figure 2). As confirmed by RT-qPCR and Western blot, E6 and P16 expression were increased in E6 overexpression HNSCC cells (Figure 2A and B) and decreased in E6 knockdown HNSCC cells (Figure 2D and E). FaDu and UM-SCC-4 cells, transfected with E6 overexpression plasmid, possessed increased clonogenic survival upon radiation (Figure 2C). On contrary, Silenced E6 in UM-SCC-47 and UPCI-SCC-090 cells showed significantly reduced clonogenic survival upon radiation compared with control cells (Figure 2F). Based on the above results, we speculated that the expression of E6 enforced the radiation sensitivity of HNSCC cells.
E6 Downregulates SMG1 Expression via DNMT1
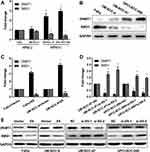
Aberrant DNA promoter methylation is common in many cancers, including HNSCC. SMG1 is reported to have a high GC content in the promoter region, which might be regulated by DNA methyltransferase 1 (DNMT1). We are interested to detect the relationship between SMG1 expression and the status of E6 in HNSCC. The mRNAs and proteins expression of DNMT1 and SMG1 in HNSCC cells are displayed in Figure 3A and B. DNMT1 expression in HPV-positive cells was significantly higher than HPV-negative cells. However, SMG1 expression in HPV-positive cells was lower than HPV-negative cells. To explore the relationship among E6, DNMT1 and SMG1, we altered E6 expression in HNSCC cells. Overexpression of E6 upregulated the DNMT1 level and downregulated the SMG1 expression level while interference of E6 leads to decreased DNMT1 expression but increased SMG1 expression (Figure 3C–E). The E6 expression level and radiotherapy sensitivity of E6 were detected. Overexpression E6 expression increased the radiotherapy sensitivity in FaDu and UM-SCC-4 (Figure 4A and B) while silence it decreased the radiotherapy sensitivity in UM-SCC-47 and UPCI-SCC-090 (Figure 4C and D). The content of E6 is related to radiotherapy sensitivity. These results suggested that HPV-E6 promote the sensitivity of radiotherapy.
Next, we analyzed the regulatory effect of DNMT1 on SMG1. As shown in Figure 5A–C, the expression of SMG1 was down-regulated by DNMT1 overexpression plasmid and up-regulated when DNMT1 was silenced. Azacitidine is a specificity DNMT1inhibitor. DNMT1 inhibitor Azacitidine continuously decreased the expression of DNMT1 protein expression over time (Figure 5D). Rescue was used to investigate the relationship between E6, DNMT1 and SMG1 by overexpressing E6 and down-regulated the expression level of DNMT1. The results showed that SMG1 expression decreased after E6 overexpression and increased after DNMT1 down0regulated in FaDu and UM-SCC-4. What's more, E6 overexpression and DNMT1 silence at the same time resulted in SMG1 expression between E6 overexpression and DNMT1 silence (Figure 5E). The above results indicated that E6 downregulates SMG1 expression via DNMT1.
Downregulation of SMG1 in HNSCC Cells Results in Increased Radiation Sensitivity
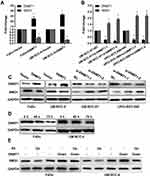
Low SMG1 expression in HPV-positive HNSCC cells might contribute to increased radiation sensitivity. To clarify this speculation, we silenced SMG1 expression in FaDu and UM-SCC-4, and assessed sensitivity of the cells to radiation in clonogenic survival assay. Both siRNAs resulted in a decreased expression of SMG1 in FaDu and UM-SCC-4 (Figure 6A and B), and showed significantly reduced clonogenic survival upon radiation compared with control (Figure 6C). The above results indicated that SMG1 increases the sensitivity of cells to radiotherapy.
Discussion
Head and neck cancers, which are variety and malignant, are the 7th malignant tumors worldwide. Previous studies have indicated that HNSCC development and radiotherapy are closed related to HPV E6/E7. The occurrence of methylation may also be related to the occurrence of tumors.21 In our study, we classify 4 HNSCC cells by detecting HPV E6/E7. FaDu and UM-SCC-4 are HPV-negative cell lines, UM-SCC-47 and UPCI-SCC-090 are HPV-positive cell lines. HPV-positive HNSCC are more sensitive to radiotherapy than HPV-negative cells. We found that E6/DNMT1/SMG1 axis affected the sensitivity of radiotherapy in HNSCC.
According to epidemiological investigations, HPV is a major agent of HNSCC.22 Over the past several decades, an increasing number of studies established the strong association of HPV with the invasion and metastasis of HNSCC. The gene mutations in HPV-associated HNSCC and the unique mechanism of E6/E7-mediated carcinogenesis via interactions with an array of cellular elements. Kindt N et al research indicated that the involvement of HPV infection in the release of macrophage migration was an inhibitory factor in HNSCC.23 Morphoproteomics studies indicated that HPV E6/E7 in-situ hybridization and biomedical analytics defined the etiopathogenesis of HPV-associated oropharyngeal carcinoma.24 Viral DNA integration and methylation of HPV were found in high-grade epithelial dysplasia and head and neck squamous cell carcinoma.25
In many clinical cases of HNSCC, mutations in the p53 have occurred.26 P53 is a tumor suppressor gene. More than 50% of all malignant tumors will have mutations in the gene. The protein encoded by p53 is a transcriptional factor that controls the initiation of the cell cycle.27 In normal conditions, many signals about cell cycle are sent to the p53.28 The protein is determined by whether or not to start cell division. If the cell is damaged and cannot be repaired, the p53 will be involved in the initiation process, causing the cell to die in apoptosis. Mutated p53 cannot effectively be monitored allowing cancer cells to proliferate rapidly.
P16, known as multiple tumor suppressor 1 (MTS1), can participate in the regulation of the cell cycle, which is closely related to tumorigenesis.29 P16 has been found to be homozygous deletion and nonsense, missense and frameshift mutation in lung cancer, breast cancer, brain tumor, bone tumor, skin cancer, bladder cancer and so on, indicating p16 are widely involved in tumor formation by deletion and mutation.30 P16 is localized in the nucleus. Chou A et al demonstrated that p16 is an inhibitor of CDK4, one of the key enzymes involved in the cell division cycle.31 It is of great clinical significance to detect the presence or absence of changes in the p16 in determining the susceptibility of a patient’s tumor and predicting the prognosis of the HNSCC.
DNMT1 is responsible for maintenance DNA methylation which ensures the fidelity of replication of inherited epigenetic patterns.32 It has a very distinguishable preference of methylating CpGs on hemimethylated DNA. Aberrant methylation patterns are associated with certain human tumors and developmental abnormalities. Au Yeung CL et al proved that HPV-16 E6 upregulation DNMT1 through repression of tumor suppressor p53.33 It has been reported that high expression of DNMT1can increased radiosensitivity.34
In this study, the expression of DNMT1 in HPV (-) cells was lower than HPV (+) cells. The possible cause of the above phenomenon was that HPV (+) cells needed more DNMT1 to maintain methylation status. The expression level of DNMT1 and methylation level are related to the expression of E6.33
For many years, SMG1 has been known as a critical component of an evolutionarily conserved mRNA surveillance mechanism.35–37 It was later discovered to be involved in the maintenance of genome integrity via genotoxic stress response pathways. SMG1 is activated not only in response to errors in mRNA processing but also in response to DNA double-strand breaks caused by IR and UV light. Effect of ionizing radiation on apoptosis and SMG1 studies have been reported.38 Gubanova E et al found the negative correlation between SMG1 and radiation sensitivity in HPV-positive HNSCC cells.3 Our study indicated that the expression of SMG1 in HPV (-) cells was higher than HPV (+) cells. The SMG1 expression decreased when overexpressing DNMT1 which inferred that the expression of SMG1 might be regulated by DNMT1 and the expression of SMG1 was one of the important factors affecting the sensitivity of radiotherapy. We demonstrated for the first time that DNMT1 down-regulates the expression of SMG1 to increase the sensitivity of radiotherapy.
In summary, this study distinguished that HPV E6 could down-regulate the expression of SMG1 by up-regulating the expression of DNMT1 in order to reduce DNA stability and improve radiotherapy. SMG1 may be an effective switch to regulate the effect of radiotherapy. By regulating the expression of SMG1, HNSCC patients can get better radiotherapy effect.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81760493 to CZ, 81860482 to ZL); The Fund from Education Department of Guizhou Province for Young scientific and Technological Talents (QKH NO. 2017-201.) Research Fund from Guizhou Provincial Health and Family Planning Commission gzwjkj2016-1-038, The PhD Start-up Fund of Zunyi Medical University (No.2015-11)
Author Contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no competing interests in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
2. Isayeva T, Li Y, Maswahu D, Brandwein-Gensler M. Human papillomavirus in non-oropharyngeal head and neck cancers: a systematic literature review. Head Neck Pathol. 2012;6(Suppl 1):S104–S120. doi:10.1007/s12105-012-0368-1
3. Gubanova E, Brown B, Ivanov SV, et al. Downregulation of SMG-1 in HPV-positive head and neck squamous cell carcinoma due to promoter hypermethylation correlates with improved survival. Clin Cancer Res. 2012;18(5):1257–1267. doi:10.1158/1078-0432.CCR-11-2058
4. Biktasova A, Gubanova E, Zou M, Yarbrough WG, Issaeva N. Abstract 5476: simultaneous targeting of multiple cancer-related pathways in HPV-positive head and neck cancer with demethylating agents. Cancer Res. 2014;74(19 Supplement):5476. doi:10.1158/0008-5472.CAN-13-3514
5. Rettig EM, D’Souza G. Epidemiology of head and neck cancer. Surg Oncol Clin N Am. 2015;24(3):379–396. doi:10.1016/j.soc.2015.03.001
6. Ou R, Zhu L, Zhao L, et al. HPV16 E7-induced upregulation of KDM2A promotes cervical cancer progression by regulating miR-132-radixin pathway. J Cell Physiol. 2019;234(3):2659–2671. doi:10.1002/jcp.v234.3
7. Rezhake R, Hu S, Zhao S, et al. Eight-type human papillomavirus E6/E7 oncoprotein detection as a novel and promising triage strategy for managing HPV-positive women. Int J Cancer. 2019;144(1):34–42. doi:10.1002/ijc.v144.1
8. de Oliveira T, Do Amaral C, de França São Marcos B, et al. Presence and activity of HPV in primary lung cancer. J Cancer Res Clin Oncol. 2018;144(12):2367–2376. doi:10.1007/s00432-018-2748-8
9. Fudulu A, Albulescu A, Anton G. Human papillomaviruses’ proteins with clinical utility. J Immunoassay Immunochem. 2018;40(1):1–10.
10. Sannigrahi M, Sharma R, Singh V, Panda N, Rattan V, Khullar M. DNA methylation regulated microRNAs in HPV-16-induced head and neck squamous cell carcinoma (HNSCC). Mol Cell Biochem. 2018;448(1–2):321–333. doi:10.1007/s11010-018-3336-6
11. Sharma S, Wagner S, Reder H, et al. The 8th edition AJCC/UICC TNM staging for p16-positive oropharyngeal carcinoma: is there space for improvement? Eur Arch Otorhinolaryngol. 2018;275(12):3087–3091. doi:10.1007/s00405-018-5156-4
12. Solomon B, Young R, Rischin D. Head and neck squamous cell carcinoma: genomics and emerging biomarkers for immunomodulatory cancer treatments. Semin Cancer Biol. 2018;52(Pt 2):228–240. doi:10.1016/j.semcancer.2018.01.008
13. Squarzanti D, Sorrentino R, Landini M, et al. Human papillomavirus type 16 E6 and E7 oncoproteins interact with the nuclear p53-binding protein 1 in an in vitro reconstructed 3D epithelium: new insights for the virus-induced DNA damage response. Virol J. 2018;15(1):176. doi:10.1186/s12985-018-1086-4
14. Deng W, Tsao S, Guan X, Cheung A. Pericentromeric regions are refractory to prompt repair after replication stress-induced breakage in HPV16 E6E7-expressing epithelial cells. PLoS One. 2012;7(10):e48576. doi:10.1371/journal.pone.0048576
15. Augustin J, Mandavit M, Outh-Gauer S, et al. HPV RNA CISH score identifies two prognostic groups in a p16 positive oropharyngeal squamous cell carcinoma population. Mod Pathol. 2018;31(11):1645–1652. doi:10.1038/s41379-018-0090-y
16. Liu H, Chen G, Vlantis A, Tong M, Chan P, van Hasselt C. Induction of cell cycle arrest and apoptosis by 5-fluorouracil in laryngeal cancer cells containing HPV16 E6 and E7 oncoproteins. Clin Biochem. 2008;41(14–15):1117–1125. doi:10.1016/j.clinbiochem.2008.06.007
17. Kojima Y, Otsuki N, Kubo M, et al. Adenovirus-mediated transfer of HPV 16 E6/E7 antisense RNA combined with cisplatin inhibits cellular growth and induces apoptosis in HPV-positive head and neck cancer cells. Cancer Gene Ther. 2018;25(9–10):274–283. doi:10.1038/s41417-018-0024-3
18. Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet. 2018;19(2):81–92. doi:10.1038/nrg.2017.80
19. Chen C, Chen W, Wang W, et al. Role of DNA methyltransferase 1 in pharyngeal cancer related to treatment resistance. Head Neck. 2011;33(8):1132–1143. doi:10.1002/hed.v33.8
20. Azzalin C, Lingner J. The double life of UPF1 in RNA and DNA stability pathways. Cell Cycle. 2006;5(14):1496–1498. doi:10.4161/cc.5.14.3093
21. Ambatipudi S, Langdon R, Richmond R, et al. DNA methylation derived systemic inflammation indices are associated with head and neck cancer development and survival. Oral Oncol. 2018;85:87–94. doi:10.1016/j.oraloncology.2018.08.021
22. Schneider K, Grønhøj C, Hahn C, von Buchwald C. Therapeutic human papillomavirus vaccines in head and neck cancer: a systematic review of current clinical trials. Vaccine. 2018;36(45):6594–6605. doi:10.1016/j.vaccine.2018.09.027
23. Kindt N, Descamps G, Lechien J, et al. Involvement of HPV infection in the release of macrophage migration inhibitory factor in head and neck squamous cell carcinoma. J Clin Med. 2019;8(1):75. doi:10.3390/jcm8010075
24. Brown R, Naqvi S, McGuire M, Buryanek J, Karni R. Morphoproteomics, E6/E7 in-situ hybridization, and biomedical analytics define the etiopathogenesis of HPV-associated oropharyngeal carcinoma and provide targeted therapeutic options. J Otolaryngol Head Neck Surg. 2017;46(1):52. doi:10.1186/s40463-017-0230-2
25. Khanal S, Shumway B, Zahin M, et al. Viral DNA integration and methylation of human papillomavirus type 16 in high-grade oral epithelial dysplasia and head and neck squamous cell carcinoma. Oncotarget. 2018;9(54):30419–30433. doi:10.18632/oncotarget.v9i54
26. Daher T, Tur M, Brobeil A, et al. Combined human papillomavirus typing and TP53 mutation analysis in distinguishing second primary tumors from lung metastases in patients with head and neck squamous cell carcinoma. Head Neck. 2018;40(6):1109–1119. doi:10.1002/hed.25041
27. Kalan S, Amat R, Schachter M, et al. Activation of the p53 transcriptional program sensitizes cancer cells to Cdk7 inhibitors. Cell Rep. 2017;21(2):467–481. doi:10.1016/j.celrep.2017.09.056
28. Fischer M, Steiner L, Engeland K. The transcription factor p53: not a repressor, solely an activator. Cell Cycle. 2014;13(19):3037–3058. doi:10.4161/15384101.2014.949083
29. Ameri A, Alidoosti A, Hosseini S, et al. Prognostic value of promoter hypermethylation of Retinoic Acid Receptor Beta (RARB) and CDKN2 (p16/MTS1) in prostate cancer. Chin J Cancer Res. 2011;23(4):306–311. doi:10.1007/s11670-011-0306-x
30. Sand F, Nielsen D, Frederiksen M, Rasmussen C, Kjaer S. The prognostic value of p16 and p53 expression for survival after vulvar cancer: a systematic review and meta-analysis. Gynecol Oncol. 2018;152(1):208–217.
31. Chou A, Froio D, Nagrial A, et al. Tailored first-line and second-line CDK4-targeting treatment combinations in mouse models of pancreatic cancer. Gut. 2018;67(12):2142–2155. doi:10.1136/gutjnl-2017-315144
32. Gowher H, Jeltsch A. Mammalian DNA methyltransferases: new discoveries and open questions. Biochem Soc Trans. 2018;46(5):1191–1202. doi:10.1042/BST20170574
33. Yeung A, Lam C, Tsang WP, et al. HPV-16 E6 upregulation of DNMT1 through repression of tumor suppressor p53. Oncol Rep. 2010;24(6):1599–1604. doi:10.3892/or_00001023
34. Terrazzino S, Deantonio L, Cargnin S, et al. DNA Methyltransferase gene polymorphisms for prediction of radiation-induced skin fibrosis after treatment of breast cancer: a multifactorial genetic approach. Cancer Res Treat. 2017;49(2):464–472. doi:10.4143/crt.2016.256
35. Ahn S, Kim J, Hwang J. CK2-mediated TEL2 phosphorylation augments nonsense-mediated mRNA decay (NMD) by increase of SMG1 stability. Biochim Biophys Acta. 2013;1829(10):1047–1055. doi:10.1016/j.bbagrm.2013.06.002
36. Brumbaugh K, Otterness D, Geisen C, et al. The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol Cell. 2004;14(5):585–598. doi:10.1016/j.molcel.2004.05.005
37. Grimson A, O’Connor S, Newman C, Anderson P. SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA Decay in Caenorhabditis elegans. Mol Cell Biol. 2004;24(17):7483–7490. doi:10.1128/MCB.24.17.7483-7490.2004
38. Chen J, Crutchley J, Zhang D, Owzar K, Kastan MB. Identification of a DNA damage-induced alternative splicing pathway that regulates p53 and cellular senescence markers. Cancer Discov. 2017;7(7):766–781. doi:10.1158/2159-8290.CD-16-0908
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.