Back to Journals » Cancer Management and Research » Volume 10
Distinct prognostic values of mRNA expression of glutathione peroxidases in non–small cell lung cancer
Authors Liu K, Jin M, Xiao L, Liu H, Wei S
Received 22 January 2018
Accepted for publication 7 May 2018
Published 28 August 2018 Volume 2018:10 Pages 2997—3005
DOI https://doi.org/10.2147/CMAR.S163432
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Antonella D'Anneo
Kui Liu, Meng Jin, Li Xiao, Huiguo Liu, Shuang Wei
Department of Respiratory and Critical Care Medicine, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430030, China
Introduction: Glutathione peroxidases (GPxs) constitutes an enzyme family which has the ability to reduce free hydrogen peroxide to water and lipid hydroperoxides to their corresponding alcohols, and its main biological roles are to protect organisms from oxidative stress-induced damage. GPxs include eight members in different tissues of the body, and they play essential roles in carcinogenesis. However, the prognostic value of individual GPx in non-small cell lung cancer (NSCLC) remains elusive.
Materials and methods: In the current study, we investigated the prognostic value of GPxs in NSCLC patients through the “Kaplan–Meier plotter” database, wherein updated gene expression data and survival information from a total of 1,926 NSCLC patients are included.
Results: High expression of GPx1 mRNA was correlated with worse overall survival (OS) in adenocarcinoma patients. High expression of GPx2 mRNA was correlated with worse OS for all NSCLC patients. In contrast, high expression of GPx3 mRNA was associated with better OS for all NSCLC patients. High expression of GPx4 mRNA was significantly correlated with worsening adenocarcinoma in these patients. GPx5 mRNA high expression correlated with worsening OS for all NSCLC patients.
Discussion: The current findings of prognostic values of individual mRNA expression of GPxs in NSCLC patients indicate some GPxs may have prognostic value in NSCLC patients, and this needs further study.
Keywords: NSCLC, glutathione peroxidases (GPxs), cancer stem cell, prognosis, KM plotter, hazard ratio
Introduction
Lung cancer remains the leading cause of cancer-related mortality worldwide in both men and women.1,2 Non–small cell lung cancer (NSCLC) includes adenocarcinoma (ADE) and squamous cell carcinoma (SCC), and accounts for 75–80% of cases of lung cancer. A multidisciplinary approach combining the application of chemotherapy with alternative treatment modalities as well as targeted therapy have aided improvement in survival outcomes. However, success with the above treatment modalities has continually been constrained by various limitations.3,4 Therefore, further investigation on the mechanism of progression, as well as identification of prognostic markers, will help select patients with a higher chance of lung cancer recurrence and provide better prognosis and individualized treatment.
Glutathione peroxidases (GPxs) belong to an enzyme family which has the ability to reduce free hydrogen peroxide to water and lipid hydroperoxides to their corresponding alcohols; their main biological roles are to protect organisms from oxidative stress-induced damage.5–8 GPxs include eight members in different tissues of the body with different substrate specificity, and they play essential roles in the protection of the organism from oxidative damage and carcinogenesis.8–10 Reactive oxygen species (ROS)-induced oxidative stress from mitochondrial dysfunction or NADPH oxidase (NOX) overactivation and ectopic expression of antioxidative enzymes were involved in EGFR-mediated tumor progression and drug resistance in malignancies including NSCLC.11,12 However, the prognostic values of individual GPxs in NSCLC remain elusive. The “Kaplan–Meier plotter” (KM plotter) generated data from the Gene Expression Omnibus (GEO, www.ncbi.nlm.nih.gov/geo/)13 database. The KM plotter database was initially established using data on expression manifested by 22,277 genes in a group of 1,809 patients with breast cancer.14,15 Later, this database also included gene expression data and survival information from a total of 1,715 NSCLC patients.16 Recently, this database has included gene expression data and survival information from patients with ovarian cancer,17 gastric cancer, as well as liver cancer.18 Thus, KM plotter can be utilized for the analysis of individual genes with clinical results to relapse-free survival and total survival of the patients. So far, a number of genes have been identified and/or validated by KM plotter in breast cancer19–27 as well as in NSCLC.28 In this study, we determined the prognostic value of individual GPx in human NSCLC patients using the KM plotter database.
Materials and methods
We used an online database14 to determine the relevance of mRNA expression of individual GPx members to relapse-free survival. The database was established using gene expression data and survival information of 1,926 NSCLC patients downloaded from Gene Expression Omnibus (GEO). Briefly, eight GPx sub-members (GPx1, GPx2, GPx3, GPx4, GPx5, GPx6, GPx7, and GPx8) were entered into the database (http://kmplot.com/analysis/index.php?p=service&cancer=lung) to obtain Kaplan–Meier survival plots in which the number-at-risk is indicated below the main plot. The certain gene mRNA expression above or below the median separates the cases into high and low expression, respectively. HR (95% CIs) and log rank p were calculated and displayed on the webpage.
Results
The GPxs family comprises a total of eight sub-members. Among all the eight GPxs isoenzymes, only GPx8 is not included in the database; GPx6 and GPx7 share the same mRNA in www.kmplot.com.
We first examined the prognostic value of GPx1 mRNA expression in www.kmplot.com. The desired Affymetrix IDs is valid: 200736_s_at (GPx1). Survival curves were plotted for all patients (n=1,926; Figure 1A), for ADE (n=720; Figure 1B), and for SCC (n=524; Figure 1C). High expression of GPx1 mRNA was not found to be correlated with overall survival (OS) for all NSCLC patients followed for 20 years (HR 1.03 [95% CI 0.9–1.16]; p=0.7). However, high expression of GPx1 mRNA was found to be correlated to worsen OS in ADE patients (HR 1.49 [95% CI 1.18–1.89]; p=0.00083), but not in SCC patients (HR 0.81 [95% CI 0.64–1.03]; p=0.082).
We then examined the prognostic value of GPx2 mRNA expression in www.kmplot.com. The desired Affymetrix ID is valid: 239595_at (GPx2). High expression of GPx2 mRNA correlated with worse OS for all NSCLC patients (HR 1.63 [95% CI 1.38–1.93]; p=5.1e-09; Figure 2A). However, high expression of GPx2 mRNA was not significantly correlated with worse OS in ADE patients (HR 1.21 [95% CI 0.95–1.54]; p=0.12; Figure 2B) and SCC patients (HR 1.37 [95% CI 1–1.87]; p=0.051; Figure 2C).
Figure 3 shows the prognostic value of GPx3 mRNA expression in www.kmplot.com. The desired Affymetrix ID is valid: 201348_at (GPx3). High expression of GPx3 mRNA correlated with better OS for all NSCLC patients (HR 0.76 [95% CI 0.67–0.86]; p=2e-05; Figure 3A). However, high expression of GPx3 mRNA was not significantly correlated with better OS in ADE patients (HR 0.85 [95% CI 0.67–1.07]; p=0.16; Figure 3B) and SCC patients (HR 0.87 [95% CI 0.69–1.11]; p=0.27; Figure 3C).
Figure 4 shows the prognostic value of GPx4 mRNA expression in www.kmplot.com. The desired Affymetrix ID is valid: 201106_at (GPx4). The curves show that high expression of GPx4 mRNA above or below the median do not separate the cases into significantly different prognostic groups among all NSCLC patients (HR 0.97 [95% CI 0.85–1.1]; p=0.63; Figure 4A). However, high expression of GPx4 mRNA was significantly correlated with worse OS in ADE patients (HR 1.3 [95% CI 1.03–1.65]; p=0.029; Figure 2B), but not in SCC patients (HR 0.98 [95% CI 0.78–1.25]; p=0.9; Figure 4C).
Figure 5 shows the prognostic value of GPx5 mRNA expression in www.kmplot.com. The desired Affymetrix ID is valid: 214648_at (GPx5). GPx5 mRNA high expression was found to be correlated with worse OS for all NSCLC patients (HR 1.19 [95% CI 1.05–1.35]; p=0.0064; Figure 5A). In addition, high expression of GPx5 mRNA was found to be correlated with worse OS in ADE patients (HR 1.57 [95% CI 1.24–1.99]; p=0.00013; Figure 5B), but not in SCC patients (HR 1.08 [95% CI 0.85–1.37]; p=0.52; Figure 5C).
Figure 6 shows the prognostic value of GPx6 or GPx7 mRNA expression in www.kmplot.com. The desired Affymetrix ID is valid: 213170_at (GPx6 or7). The curves show that high expression of GPx6/7 mRNA above or below the median do not separate the cases into significantly different prognostic groups in all NSCLC patients (HR 1.13 [95% CI 1–1.28]; p=0.056; Figure 6A), in ADE patients (HR 1.19 [95% CI 0.94–1.5]; p=0.15; Figure 6B), as well as in SCC patients (HR 0.95 [95% CI 0.75–1.2]; p=0.67; Figure 6C).
To further determine the correlation of individual GPx with other clinicopathological features, we determined their correlation with the smoking status (Table 1), clinical stages (Table 2), and chemotherapy (Table 3) of NSCLC patients. As shown in Table 1, high expression of GPx5, GPx6, or GPx7 mRNA was correlated with worse OS in never-smoked NSCLC patients. From Table 2, it is apparent that GPx2, GPx3, GPx6, or GPx7 are significantly associated with clinical stages of NSCLC patients. From the data in Table 3, it is evident that none of the GPxs are significantly associated with NSCLC patients, with or without chemotherapy, probably due to the relatively limited number of patients.
  | Table 1 Correlation of GPx mRNA expression with smoking status of NSCLC patients Abbreviations: GPxs, glutathione peroxidases; NSCLC, non-small cell lung cancer. |
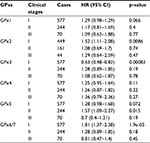  | Table 2 Correlation of GPx mRNA expression with clinical stages of NSCLC patients Abbreviations: GPxs, glutathione peroxidases; NSCLC, non-small cell lung cancer. |
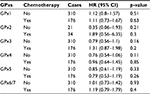  | Table 3 Correlation of GPx mRNA expression with chemotherapy of NSCLC patients Abbreviations: GPxs, glutathione peroxidases; NSCLC, non-small cell lung cancer. |
Discussion
Changes in GPx levels in several types of tumor have been reported. GPx1 was reported to prevent oxidative DNA mutations and, thus, GPx1 may prevent tumorigenesis.29 Overexpressed GPx1 reduced tumor growth, which indicates that it has a role in protecting against tumorigenesis.30 The dual role of GPx2 in tumorigenesis has been reviewed recently.9,31 Overexpression of GPx2 was observed in several tumors,32,33 including lung cancer,34 indicating that GPx2 may be an oncogene. GPx3 is considered to be a novel tumor suppressor, as hypermethylation of GPx3 was detected in tumor samples from patients with Barrett’s esophagus35–37 as well as in endometrial38 and prostate cancers,39 and downregulation of GPx3 was generally correlated with worse prognosis. Moreover, GPx4 is considered to be a tumor suppressor, because it is downregulated in pancreatic30 and breast cancers.40 In addition, GPx4 overexpression reduced fibrosarcoma cell growth.41 So far, there are no reports on the role of GPx5, -6, -7, and -8 in tumorigenesis. With the exception of GPx3, all GPx members were reported to be associated with the prognosis of tumor patients. For example, increased GPx1 expression was significantly associated with poor prognosis in oral SCC42 and laryngeal SCC.43 In contrast, loss of GPx1 expression was associated with aggressiveness and poor survival in patients with gastric cancer.43 Furthermore, GPx2 overexpression was correlated with poor prognosis in patients with hepatocellular carcinoma45 and gastric cancer.46 In contrast, loss of GPx2 expression was associated with patients who had esophageal SCC.47 The distinct different prognostic values of GPx1 and GPx2 in different types of cancer patients have not been well explained so far.48
Several reports exist about the liver changes associated with mRNA expression of GPxs in NSCLC cell lines and/or tumor tissues;49–51 however, the prognostic value of mRNA expression of GPxs in NSCLC patients has not been reported. KM plotter is a widely used database containing gene expression data and survival information.17 Until now, a number of genes, such as ALDH1, GLI1, ITIH5, CK2, RAI14, and GREB1, have been identified and validated by KM plotter in lung,16,28,52,53 breast,19–28,54 gastric,55,56 and ovarian cancers.28,57,58 Using KM plotter, we determined the prognostic value of individual GPx mRNA in NSCLC patients. High expression of GPx1 mRNA was not correlated with OS for all NSCLC patients followed for 20 years. However, high expression of GPx1 mRNA was correlated with worse OS in ADE patients (HR 1.49 [95% CI 1.18–1.89]; p=0.00083). High expression of GPx2 mRNA correlated with worse OS for all NSCLC patients (HR 1.63 [95% CI 1.38–1.93]; p=5.1e-09). In contrast, high expression of GPx3 mRNA correlated with better OS for all NSCLC patients (HR 0.76 [95% CI 0.67–0.86]; p=2e-05). High expression of GPx4 mRNA was significantly correlated with worse OS in ADE patients (HR 1.3 [95% CI 1.03–1.65]; p=0.029). High expression of GPx5 mRNA correlated with worse OS for all NSCLC patients (HR 1.19 [95% CI 1.05–1.35]; p=0.0064). High expression of GPx6/7mRNA was not correlated with OS for all NSCLC patients. GPx8, as a novel member belonging to the GPx family, has been identified in a phylogenetic analysis in amphibia and mammalia.59 GPx8 is a membrane protein, lung-abundant enzyme, and is detected in the endoplasmic reticulum.60,61 However, little is known about its role. In addition, individual GPx may interact among themselves and finally affect the prognosis of NSCLC patients. However, the KM plotter cannot be used to analysis the impact between the various isoforms of GPx. Moreover, the KM plotter cannot be used to analyze the correlation between isoenzyme expression.
Nicotine can regulate a number of biological functions such as cell proliferation, invasion, inflammation, apoptosis, and angiogenesis.62 Through inducing the secretion of stem cell factors, nicotine is able to regulate the growth and metastasis of NSCLC.63 There is no evidence about the direct correlation between nicotine and GPx expression in NSCLC. However, nicotine treatment may significantly impact the expression of GPx1 in peripheral blood lymphocytes,64 rat kidneys,65 as well as in a novel cell line – Danio rerio gill (DrG) – derived from the gill tissue of zebrafish.66 Furthermore, nicotine treatment was reported to significantly impact the expression of GPx4, GPx6, and GPx7 in rat brain.67 In this report, we observed that high expression of GPx5, GPx6, or GPx7 mRNA correlated with worse OS in never-smoked NSCLC patients.
There are a number of reports about the prognostic values of GPxs in tumor patients. For example, loss of GPx1 expression was associated with tumor aggressiveness and poor survival in patients with gastric cancer.43 GPx2 overexpression indicates poor prognosis in patients with hepatocellular carcinoma,45 gastric cancer,46 and urothelial carcinomas of the upper urinary tract and urinary bladder.68 However, patients with prostate cancer with high GPx2 expression on the biopsy specimen had significantly lower prostate-specific antigen (PSA), recurrence-free survival, and OS than those without any GPx2 expression.69 Downregulation of GPx3 significantly correlated with poor prognosis in hepatocellular carcinoma,70 cervical cancer,71 and gallbladder cancer.72 GPx3 is considered to be a novel tumor suppressor, because hypermethylation of the GPx3 promoter was detected in tumor samples from patients with Barrett’s esophagus35–37 as well as endometrial38 and prostate cancers,39 and downregulation of GPx3 was generally associated with a poor prognosis. However, so far, no prognostic significance of GPxs protein expression in NSCLC patients has been reported. The current findings of prognostic values of individual mRNA expression of GPxs in NSCLC patients indicate some members of the GPx family may also have prognostic values in NSCLC patients, and this needs further study.
Acknowledgment
The work was supported by the National Natural Science Foundation of China (grant no. 81300064).
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. | ||
Guo P, Huang ZL, Yu P, Li K. Trends in cancer mortality in China: an update. Ann Oncol. 2012;23(10):2755–2762. | ||
Lazzari C, Karachaliou N, Bulotta A, et al. Combination of immunotherapy with chemotherapy and radiotherapy in lung cancer: is this the beginning of the end for cancer? Ther Adv Med Oncol. 2018;10:1758835918762094. | ||
Jones CM, Brunelli A, Callister ME, Franks KN. Multimodality treatment of advanced non-small cell lung cancer: where are we with the evidence? Curr Surg Rep. 2018;6(2):5. | ||
Lubos E, Loscalzo J, Handy DE. Glutathione peroxidase-1 in health and disease: from molecular mechanisms to therapeutic opportunities. Antioxid Redox Signal. 2011;15(7):1957–1997. | ||
Battin EE, Brumaghim JL. Antioxidant activity of sulfur and selenium: a review of reactive oxygen species scavenging, glutathione peroxidase, and metal-binding antioxidant mechanisms. Cell Biochem Biophys. 2009;55(1):1–23. | ||
Toppo S, Flohé L, Ursini F, Vanin S, Maiorino M. Catalytic mechanisms and specificities of glutathione peroxidases: variations of a basic scheme. Biochim Biophys Acta. 2009;1790(11):1486–1500. | ||
Brigelius-Flohé R, Kipp A. Glutathione peroxidases in different stages of carcinogenesis. Biochim Biophys Acta. 2009;1790(11):1555–1568. | ||
Brigelius-Flohé R, Kipp AP. Physiological functions of GPx2 and its role in inflammation-triggered carcinogenesis. Ann N Y Acad Sci. 2012;1259:19–25. | ||
Zhuo P, Diamond AM. Molecular mechanisms by which selenoproteins affect cancer risk and progression. Biochim Biophys Acta. 2009;1790(11):1546–1554. | ||
Weng MS, Chang JH, Hung WY, Yang YC, Chien MH. The interplay of reactive oxygen species and the epidermal growth factor receptor in tumor progression and drug resistance. J Exp Clin Cancer Res. 2018;37(1):61. | ||
Kumari S, Badana AK, G MM, G S, Malla R. Reactive oxygen species: a key constituent in cancer survival. Biomark Insights. 2018;13:1177271918755391. | ||
Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002 Jan 1;30(1):207–210. | ||
Györffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123(3):725–731. | ||
Györffy B, Benke Z, Lánczky A, et al. RecurrenceOnline: an online analysis tool to determine breast cancer recurrence and hormone receptor status using microarray data. Breast Cancer Res Treat. 2012;132(3):1025–1034. | ||
Györffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8(12):e82241. | ||
Gyorffy B, Lánczky A, Szállási Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr Relat Cancer. 2012;19(2):197–208. | ||
Szász AM, Lánczky A, Nagy Á, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7(31):49322–49333. | ||
Liu M, Wang G, Gomez-Fernandez CR, Guo S. GREB1 functions as a growth promoter and is modulated by IL6/STAT3 in breast cancer. PLoS One. 2012;7(10):e46410. | ||
Tilghman SL, Townley I, Zhong Q, et al. Proteomic signatures of acquired letrozole resistance in breast cancer: suppressed estrogen signaling and increased cell motility and invasiveness. Mol Cell Proteomics. 2013;12(9):2440–2455. | ||
Zhou C, Zhong Q, Rhodes LV, et al. Proteomic analysis of acquired tamoxifen resistance in MCF-7 cells reveals expression signatures associated with enhanced migration. Breast Cancer Res. 2012; 14(2):R45. | ||
Maciejczyk A, Szelachowska J, Czapiga B, et al. Elevated BUBR1 expression is associated with poor survival in early breast cancer patients: 15-year follow-up analysis. J Histochem Cytochem. 2013;61(5):330–339. | ||
Maciejczyk A, Łacko A, Ekiert M, et al. Elevated nuclear S100P expression is associated with poor survival in early breast cancer patients. Histol Histopathol. 2013;28(4):513–524. | ||
Maciejczyk A, Jagoda E, Wysocka T, et al. ABCC2 (MRP2, cMOAT) localized in the nuclear envelope of breast carcinoma cells correlates with poor clinical outcome. Pathol Oncol Res. 2012;18(2):331–342. | ||
Adam MA. New prognostic factors in breast cancer. Adv Clin Exp Med. 2013;22(1):5–15. | ||
Ivanova L, Zandberga E, Silina K, et al. Prognostic relevance of carbonic anhydrase IX expression is distinct in various subtypes of breast cancer and its silencing suppresses self-renewal capacity of breast cancer cells. Cancer Chemother Pharmacol. 2015;75(2):235–246. | ||
Wu S, Xue W, Huang X, et al. Distinct prognostic values of ALDH1 isoenzymes in breast cancer. Tumour Biol. 2015;13:13. | ||
Ortega CE, Seidner Y, Dominguez I. Mining CK2 in cancer. PLoS One. 2014;9(12):e115609. | ||
Baliga MS, Wang H, Zhuo P, Schwartz JL, Diamond AM. Selenium and GPx-1 overexpression protect mammalian cells against UV-induced DNA damage. Biol Trace Elem Res. 2007;115(3):227–242. | ||
Liu J, Du J, Zhang Y, et al. Suppression of the malignant phenotype in pancreatic cancer by overexpression of phospholipid hydroperoxide glutathione peroxidase. Hum Gene Ther. 2006;17(1):105–116. | ||
Brigelius-Flohé R, Maiorino M. Glutathione peroxidases. Biochim Biophys Acta. 2013;1830(5):3289–3303. | ||
Al-Taie OH, Uceyler N, Eubner U, et al. Expression profiling and genetic alterations of the selenoproteins GI-GPx and SePP in colorectal carcinogenesis. Nutr Cancer. 2004;48(1):6–14. | ||
Chiu ST, Hsieh FJ, Chen SW, Chen CL, Shu HF, Li H. Clinicopathologic correlation of up-regulated genes identified using cDNA microarray and real-time reverse transcription-PCR in human colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2005;14(2):437–443. | ||
Woenckhaus M, Klein-Hitpass L, Grepmeier U, et al. Smoking and cancer-related gene expression in bronchial epithelium and non-small-cell lung cancers. J Pathol. 2006;210(2):192–204. | ||
Lee OJ, Schneider-Stock R, McChesney PA, et al. Hypermethylation and loss of expression of glutathione peroxidase-3 in Barrett’s tumorigenesis. Neoplasia. 2005;7(9):854–861. | ||
Peng DF, Razvi M, Chen H, et al. DNA hypermethylation regulates the expression of members of the Mu-class glutathione S-transferases and glutathione peroxidases in Barrett’s adenocarcinoma. Gut. 2009;58(1):5–15. | ||
He Y, Wang Y, Li P, Zhu S, Wang J, Zhang S. Identification of GPX3 epigenetically silenced by CpG methylation in human esophageal squamous cell carcinoma. Dig Dis Sci. 2011;56(3):681–688. | ||
Falck E, Karlsson S, Carlsson J, Helenius G, Karlsson M, Klinga-Levan K. Loss of glutathione peroxidase 3 expression is correlated with epigenetic mechanisms in endometrial adenocarcinoma. Cancer Cell Int. 2010;10:46. | ||
Yu YP, Yu G, Tseng G, et al. Glutathione peroxidase 3, deleted or methylated in prostate cancer, suppresses prostate cancer growth and metastasis. Cancer Res. 2007;67(17):8043–8050. | ||
Cejas P, García-Cabezas MA, Casado E, et al. Phospholipid hydroperoxide glutathione peroxidase (PHGPx) expression is downregulated in poorly differentiated breast invasive ductal carcinoma. Free Radic Res. 2007;41(6):681–687. | ||
Heirman I, Ginneberge D, Brigelius-Flohé R, et al. Blocking tumor cell eicosanoid synthesis by GP x 4 impedes tumor growth and malignancy. Free Radic Biol Med. 2006;40(2):285–294. | ||
Lee JR, Roh JL, Lee SM, et al. Overexpression of glutathione peroxidase 1 predicts poor prognosis in oral squamous cell carcinoma. J Cancer Res Clin Oncol. 2017;143(11):2257–2265. | ||
Zhang Q, Xu H, You Y, Zhang J, Chen R. High Gpx1 expression predicts poor survival in laryngeal squamous cell carcinoma. Auris Nasus Larynx. 2018;45(1):13–19. | ||
Min SY, Kim HS, Jung EJ, Jung EJ, Jee CD, Kim WH. Prognostic significance of glutathione peroxidase 1 (GPX1) down-regulation and correlation with aberrant promoter methylation in human gastric cancer. Anticancer Res. 2012;32(8):3169–3175. | ||
Liu T, Kan XF, Ma C, et al. GPX2 overexpression indicates poor prognosis in patients with hepatocellular carcinoma. Tumour Biol. 2017;39(6):1010428317700410. | ||
Liu D, Sun L, Tong J, Chen X, Li H, Zhang Q. Prognostic significance of glutathione peroxidase 2 in gastric carcinoma. Tumour Biol. 2017;39(6):1010428317701443. | ||
Lei Z, Tian D, Zhang C, Zhao S, Su M. Clinicopathological and prognostic significance of GPX2 protein expression in esophageal squamous cell carcinoma. BMC Cancer. 2016;16:410. | ||
Jiao Y, Wang Y, Guo S, Wang G. Glutathione peroxidases as oncotargets. Oncotarget. 2017;8(45):80093–80102. | ||
Gresner P, Gromadzinska J, Jablonska E, Kaczmarski J, Wasowicz W. Expression of selenoprotein-coding genes SEPP1, SEP15 and hGPX1 in non-small cell lung cancer. Lung Cancer. 2009;65(1):34–40. | ||
Hu Y, Rahlfs S, Mersch-Sundermann V, Becker K. Resveratrol modulates mRNA transcripts of genes related to redox metabolism and cell proliferation in non-small-cell lung carcinoma cells. Biol Chem. 2007;388(2):207–219. | ||
Romanowska M, Kikawa KD, Fields JR, et al. Effects of selenium supplementation on expression of glutathione peroxidase isoforms in cultured human lung adenocarcinoma cell lines. Lung Cancer. 2007;55(1):35–42. | ||
You Q, Guo H, Xu D. Distinct prognostic values and potential drug targets of ALDH1 isoenzymes in non-small-cell lung cancer. Drug Des Devel Ther. 2015;9:5087–5097. | ||
Dötsch MM, Kloten V, Schlensog M, et al. Low expression of ITIH5 in adenocarcinoma of the lung is associated with unfavorable patients’ outcome. Epigenetics. 2015;10(10):903–912. | ||
Hong CQ, Zhang F, You YJ, et al. Elevated C1orf63 expression is correlated with CDK10 and predicts better outcome for advanced breast cancers: a retrospective study. BMC Cancer. 2015;15:548. | ||
Yu T, Jia W, An Q, Cao X, Xiao G. Bioinformatic analysis of GLI1 and related signaling pathways in chemosensitivity of gastric cancer. Med Sci Monit. 2018;24:1847–1855. | ||
He XY, Zhao J, Chen ZQ, Jin R, Liu CY. High expression of retinoic acid induced 14 (RAI14) in gastric cancer and its prognostic value. Med Sci Monit. 2018;24:2244–2251. | ||
Kamieniak MM, Rico D, Milne RL, et al. Deletion at 6q24.2-26 predicts longer survival of high-grade serous epithelial ovarian cancer patients. Mol Oncol. 2015;9(2):422–436. | ||
Gayarre J, Kamieniak MM, Cazorla-Jiménez A, et al. The NER-related gene GTF2H5 predicts survival in high-grade serous ovarian cancer patients. J Gynecol Oncol. 2016;27(1):e7. | ||
Toppo S, Vanin S, Bosello V, Tosatto SC. Evolutionary and structural insights into the multifaceted glutathione peroxidase (Gpx) superfamily. Antioxid Redox Signal. 2008;10(9):1501–1514. | ||
Nguyen VD, Saaranen MJ, Karala AR, et al. Two endoplasmic reticulum PDI peroxidases increase the efficiency of the use of peroxide during disulfide bond formation. J Mol Biol. 2011;406(3):503–515. | ||
Yamada Y, Limmon GV, Zheng D, et al. Major shifts in the spatio-temporal distribution of lung antioxidant enzymes during influenza pneumonia. PLoS One. 2012;7(2):e31494. | ||
Cardinale A, Nastrucci C, Cesario A, Russo P. Nicotine: specific role in angiogenesis, proliferation and apoptosis. Crit Rev Toxicol. 2012;42(1):68–89. | ||
Perumal D, Pillai S, Nguyen J, Schaal C, Coppola D, Chellappan SP. Nicotinic acetylcholine receptors induce c-Kit ligand/Stem Cell Factor and promote stemness in an ARRB1/ β-arrestin-1 dependent manner in NSCLC. Oncotarget. 2014;5(21):10486–10502. | ||
Sudheer AR, Muthukumaran S, Kalpana C, Srinivasan M, Menon VP. Protective effect of ferulic acid on nicotine-induced DNA damage and cellular changes in cultured rat peripheral blood lymphocytes: a comparison with N-acetylcysteine. Toxicol In Vitro. 2007;21(4):576–585. | ||
Akkoyun HT, Karadeniz A. Investigation of the protective effect of ellagic acid for preventing kidney injury in rats exposed to nicotine during the fetal period. Biotech Histochem. 2016;91(2):108–115. | ||
Nathiga Nambi KS, Abdul Majeed S, Taju G, Sivasubbu S, Sarath Babu V, Sahul Hameed AS. Effects of nicotine on zebrafish: a comparative response between a newly established gill cell line and whole gills. Comp Biochem Physiol C Toxicol Pharmacol. 2017;195:68–77. | ||
Song G, Nesil T, Cao J, Yang Z, Chang SL, Li MD. Nicotine mediates expression of genes related to antioxidant capacity and oxidative stress response in HIV-1 transgenic rat brain. J Neurovirol. 2016;22(1):114–124. | ||
Chang IW, Lin VC, Hung CH, et al. GPX2 underexpression indicates poor prognosis in patients with urothelial carcinomas of the upper urinary tract and urinary bladder. World J Urol. 2015;33(11):1777–1789. | ||
Naiki T, Naiki-Ito A, Asamoto M, et al. GPX2 overexpression is involved in cell proliferation and prognosis of castration-resistant prostate cancer. Carcinogenesis. 2014;35(9):1962–1967. | ||
Qi X, Ng KT, Lian QZ, et al. Clinical significance and therapeutic value of glutathione peroxidase 3 (GPx3) in hepatocellular carcinoma. Oncotarget. 2014;5(22):11103–11120. | ||
Zhang X, Zheng Z, Yingji S, et al. Downregulation of glutathione peroxidase 3 is associated with lymph node metastasis and prognosis in cervical cancer. Oncol Rep. 2014;31(6):2587–2592. | ||
Yang ZL, Yang L, Zou Q, et al. Positive ALDH1A3 and negative GPX3 expressions are biomarkers for poor prognosis of gallbladder cancer. Dis Markers. 2013;35(3):163–172. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.






