Back to Journals » OncoTargets and Therapy » Volume 13
Disruption of LTBP4 Induced Activated TGFβ1, Immunosuppression Signal and Promoted Pulmonary Metastasis in Hepatocellular Carcinoma
Authors Yang X, Ye X, Zhang L, Zhang X, Shu P
Received 21 January 2020
Accepted for publication 22 June 2020
Published 20 July 2020 Volume 2020:13 Pages 7007—7017
DOI https://doi.org/10.2147/OTT.S246766
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Xiou Yang,1 Xiaojuan Ye,2 Liuyan Zhang,3 Xingguo Zhang,4 Peng Shu4
1Department of Infectious Disease, The People’s Hospital of Beilun District, Beilun Branch Hospital of the First Affiliated Hospital of Medical School Zhejiang University, Ningbo 315800, People’s Republic of China; 2Department of Hematology and Oncology, The People’s Hospital of Beilun District, Beilun Branch Hospital of the First Affiliated Hospital of Medical School Zhejiang University, Ningbo 315800, People’s Republic of China; 3Department of Obstetrics and Gynecology, The People’s Hospital of Beilun District, Beilun Branch Hospital of the First Affiliated Hospital of Medical School Zhejiang University, Ningbo 315800, People’s Republic of China; 4Molecular Laboratory, The People’s Hospital of Beilun District, Beilun Branch Hospital of the First Affiliated Hospital of Medical School Zhejiang University, Ningbo 315800, People’s Republic of China
Correspondence: Peng Shu
Molecular Laboratory, The People’s Hospital of Beilun District, Beilun Branch Hospital of the First Affiliated Hospital of Medical School Zhejiang University, 1288 Lushan East Road, Beilun District, Ningbo 315800, People’s Republic of China
Email [email protected]
Introduction: The current prognosis of hepatocellular carcinoma (HCC) is unsatisfactory due to high rates of recurrence and metastasis, which has led to research focused on the discovery of metastasis genes.
Methods: In this study, we combined in silico analysis and in vitro transwell experiments to identify a metastasis gene. Then, we used an in vivo experiment to validate the metastasis. Furthermore, a series of experiments such as FACS, Western blot, and ELISA were applied to explore the function of the metastasis gene.
Results: LTBP4 (latent transforming growth factor beta binding protein 4) was confirmed as a metastasis gene, whose expression levels are correlated with the overall survival rate of HCC patients. We further showed that the knockout of LTBP4 in an HCC cell line increased cell proliferation, activated the cell cycle, and induced metastasis events. Moreover, we proved that LTBP4-KO could increase the percentage of active TGFβ 1 secreted by HCC cell lines, as well as the recruitment of MDSCs (myeloid-derived suppressor cells) by active TGFβ 1 (transforming growth factor beta 1), which further inhibited CD8+ T cell proliferation and activated the immune suppression signal.
Conclusion: Our results demonstrate that the LTBP4-TGFβ 1-MDSCs axis is a critical pathway for the immune suppression signals of HCC primary tumors.
Keywords: hepatocellular carcinoma, LTBP4, TGFβ 1, MDSCs, T cell proliferation
Introduction
Hepatocellular carcinoma (HCC) the fifth most common cancer in the world, ranking third in terms of cancer-related deaths.1 More than 700,00 new cases of HCC and 250,000 deaths are reported every year.2 China accounts for over 50% of new cases and deaths, which is closely related to the large incidence of hepatitis B virus infection. Other pathogenic factors in China include the consumption of alcohol and aflatoxin, among others.3 Along with the high incidence of HCC, treatment of HCC is limited to surgical interventions during the early stages of the disease or liver transplantation.4 However, only 20% of HCC patients qualify for surgery due to the high rates of intrahepatic metastasis, portal vein cancer thrombus, and distant metastasis.5 Therefore, elucidating the oncogenes or suppressor genes involved in metastasis would help to predict HCC metastasis and identify new therapeutic targets.
For the initiation of metastasis, tumor cells require a pre-metastatic niche formed by myeloid-derived suppressor cells (MDSCs), tumor-associated macrophages (TAMs), and tumor-associated neutrophils (TANs), among others.6 The interaction between tumor cells and other cells in this pre-metastatic niche is bi-directional; on one hand, tumor cells secrete tumor-derived soluble factor (TDSFs) such as LOX and LOXL2 under hypoxia conditions, then recruit MDSCs, and simulate immune suppression signal7, while on the other hand, the tumor microenvironment, such as in the extracellular matrix (ECM), secretes soluble factors including IL-18, and supports tumor progression.8 Here, we used in vitro sgRNAs screens and identified LTBP4 (latent transforming growth factor beta binding protein 4) as a novel metastatic suppressor. In addition, we found that after LTBP4 inhibition, active TGFβ1 (transforming growth factor beta 1) secreted by the tumor recruited and simulated MDSCs, thus further inhibiting the proliferation of CD8+ T cells, and promoting the metastasis ability of HCC cells.
Materials and Methods
Ethics Statement
The animal experiment performed in this study was approved by the Ethics Committee of Ningbo Beilun People’s Hospital. All of the protocols were performed in accordance with the relevant guidelines and regulations formulated by the Ningbo Beilun People’s Hospital, Ningbo in Zhejiang province, China.
In silico Data Analysis
The in silico data analysis was performed using R 3.5.2. To identify the genes down-regulated in liver cancer patients, we downloaded the GSE14520 datasets from the GEO (Gene Expression Omnibus) database using the GEOquery package. The microarray data of 445 samples, including 220 liver non-tumor tissue and 225 liver tumor tissue, were normalized using the preprocessCore package. The down-regulated genes in the liver tumor tissue were quantified using the limma package. Genes with p < 0.05 and LogFC < ‒0.6 were identified as down-regulated genes. The pathway enrichment in the down-regulated genes was then calculated using clusterProfiler package.
To determine the relationship between LTBP4 expression and the overall survival rate of patients with liver cancer, RNA-seq and clinical data of liver hepatocellular carcinoma obtained from the TCGA (The Cancer Genome Atlas Program) were downloaded from the GDAC firehose website (http://gdac.broadinstitute.org/). The overall survival rate of the patients was assessed using the survival package. Hepatocellular carcinoma patients from TCGA were also divided into low and high LTBP4 expression groups. The proportion of immune cells in the primary tumors was calculated using the CIBERSORT website (https://cibersort.stanford.edu).
Materials
The Transwell chamber was purchased from Corning company (USA). Propidium iodide (PI) for the cell cycle was purchased from Sigma Aldrich (St Louis, MO, USA). CCK8 was purchased from Dojindo Molecular Technologies (Rockville, MD, USA). For Western blotting, the antibodies p-Smad2/3, p-Smad4, N-cadherin, E-cadherin, and Vimentin were purchased from Cell Signaling (MA, USA), while the LTBP4, TGFβ1, TGFβ2, TGFβ3, ARG1, and NOS2 antibodies were purchased from Abcam (MA, USA). LAP (latency-associated peptide) antibody was purchased from Thermo Fisher Scientific (MA, USA). The total and active TGFβ1 ELISA kits were purchased from BioLegend (CA, USA). The mass cytometry (CyTOF) antibodies were purchased from Fluidigm (CA, USA). The CD11B, LY6G, and LY6C antibodies were purchased from BioLegend (CA, USA). IgG-IP beads were purchased from Sigma Aldrich (MO, USA).
Cell Culture
Human hepatocellular carcinoma cell line Hep G2 and human embryonic kidney cell line 293t were purchased from ATCC (American Type Culture Collection) company (USA). Cells were cultured in DMEM + 10% fetal bovine serum (FBS) with 37°C at 5% CO2.
Preparation of LTBP4-KO Cells
1) First, an oligo was synthesized by Beijing Genomics institution company, the sequence of the oligo was confirmed by screen data, LTBP4 sgRNA-1:5ʹ-CCCCCGGGACCTCGACGACC-3ʹ. 2) The oligo was then cloned to lentiCRISPR v2 vector (addgene 52961) by following the instruction which is described in “entiCRISPRv2 and lentiGuide oligo cloning protocol” (https://media.addgene.org/data/plasmids/52/52961/52961-attachment_B3xTwla0bkYD.pdf). 3) The LTBP4-KO vector was then packaged as lentivirus by using the 2nd generation of the package system. 4) The HCC cell line was infected with CON (without sgRNA sequence) and LTBP4-KO virus for 48 h, and selected by puromycin for 48 h. 5) The LTBP4-KO clone was then confirmed by a single clone experiment.
Wound Healing Assay
In the wound healing assay, cells were plated on 24-well plates. The plates were scratched, then imaged every 2 h.
Cell Cycle and Proliferation
In the cell cycle experiment: 1) One million cells were harvested, followed by washing with PBS three times. 2) The cells were suspended in 300 µL PBS + 5% FBS before adding 700 µL of ethanol for cell fixation. 3) The fixed cells were treated with RNAase and labeled with PI at a final concentration of 50 ng/mL, and the cell cycle was detected using BD FACS Calibur in the FL2-A channel.
In the cell proliferation assay: 1) The cells were plated in a 96-well plate, where each well was comprised of 3000 cells in 100 µL DMEM + 10% FBS. 2) Then, 10 µL of CCK8 were added to each well before culturing at 37°C and 5% CO2 for 2 h. The absorbance was detected at 450 nM using a Varioskan™ LUX multimode microplate reader (Thermo Fisher Scientific). 3) The absorbance was detected after 24, 48, 72, and 96 h, and the proliferation index was calculated as follows: proliferation index = absorbance at time point/absorbance at 0 h.
Western Blotting
First, the cells were harvested and lysed using RIPA buffer. The cells were then mixed with loading buffer and boiled at 100°C. Second, after quantifying the concentration of the proteins in each sample using Lowry’s method, equal quantities of protein were loaded onto a 10% SDS-PAGE gel and then transferred onto a PVDF membrane. Finally, the protein extract was labeled by incubating the membranes with primary antibody at 4°C overnight, followed by incubation with the corresponding secondary antibody for 30 min at room temperature. Protein expression was quantified using Thermo Pierce ECL Western Blotting Substrate.
Endogenous Immunoprecipitation (IP)
Cells were lysed using IP lysis buffer, followed by labeling with TGFβ1 antibody by incubating at 4°C overnight. Cell lysis was then labeled with IgG-IP beads. After immunoprecipitation, cell lysis was labeled with LAP antibody. LAP binding with TGFβ1 was determined by Western blotting.
Animal Experiment
In this study, nude mice, SCID mice, and C57BL/6 mice were utilized for different experiments: 1) For the in vivo metastasis experiments, nude mice were injected with one million cells via the tail vein. After 8 weeks, the mice were sacrificed and the lung metastatic nodes were quantified by fluorescence. 2) For experiments that included primary tumors, SCID mice were subcutaneously injected with five million cells. After 4 weeks, the mice were sacrificed and the primary tumors were extracted for further study. 3) To obtain MDSCs and CD8+ T cells, C57BL/6 mice were sacrificed. The bone marrow was then flushed out and the spleens were extracted for further experiments.
Total and Active TGFβ1 Detection
1) After determining the number of cells, the plates were washed 4 times and treated with 50 µL assay buffer C. 2) This was followed by the addition of 50 µL of diluted standards or samples. 3) The plates were washed another four times and 100 µL of detection antibody solution was added. 3) The plates were washed four more times and 100 µL of avidin-HRP D solution was added. 4) The plates were washed five times and 100 µL of substrate solution was added. 5) Finally, 100 µL of stop solution was added to the plates. The absorbance was read at 450 nM.
CyTOF and FACS (Fluorescence-Activated Cell Sorting)
In the CyTOF experiment, the primary tumors were digested with Type I collagen for 2 h. The cells were then filtered using a 40 µM strainer and counted. Three million cells were labeled with the CD3E, CD8A, CD11B, LY6G, and LY6C antibodies using Fluidigm mass cytometry labeling according to the manufacturer’s instructions. In the FACS experiment, mice bone marrow and spleen tissue were extracted and digested with Type I collagen. Single cells in bone marrow were labeled with CD11B, LY6G, and LY6C antibody, to sort out CD11B LY6G+ cells (PMN MDSCs) and CD11B LY6C+ (MO MDSCs). Single cells in spleen were labeled with CD3, CD8 antibody, to sort out CD3+, CD8+ T cells.
In vitro T Cell Proliferation Experiment
After obtaining T cells from the spleen and MDSCs from the bone marrow, MDSCs were treated with cell supernatant + GM-CSF for 96 h. Simultaneously, T cells were cultured on CD3/CD28 conjugated plates for 72 h, then labeled with Dye670nm using the standard labeling procedure. The resulting MDSCs and T cells were then mixed at the indicated ratio, and T cell proliferation was determined by the BD FACS Calibur machine.
Statistical Analysis
The significance levels in this study were determined using the Student’s t-test: *p<0.05, **p<0.01, ***p<0.001.
Results
In vitro Screening of Lung Metastasis Suppressor from ECM Genes (GO:0031012)
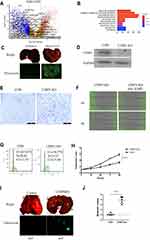
In this study, we extracted the GSE14520 dataset and found that ECM genes (GO:0031012) were highly enriched in low-expressed genes (Figure 1A). Using KEGG, we further proved that ECM-related pathways were highly enriched (Figure 1B), based on 80 ECM genes. Based on the low-expressed genes in the GSE14520 dataset, we designed 240 sgRNAs, which were transfected into Hep G2 cells using lentivirus to determine the changes in metastasis (Figure 1C). Sanger sequencing demonstrated that the resulting metastatic tumors contained 18 sgRNAs (Table 1). After extracting the sgRNA sequence and single validating using a Transwell assay, the migration ability of LTBP4-KO cells was found to be highly increased (Figure 1D and E). Furthermore, the in vitro experimental results also indicated that the blockage of TGFβ1 and its receptor inhibited cell migration in LTBP4-KO cells (Figure 1F). LTBP4-KO activated the HCC cell cycle (Figure 1G) and cell proliferation (Figure 1H). Furthermore, LTBP4-KO increased the metastasis ability of HCC cells (Figure 1I and J).
 |
Table 1 Detected sgRNAs from Metastasis Tumor of Lung |
Lower Expression of LTBP4 in HCC Tissue
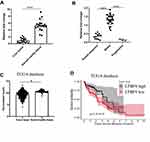
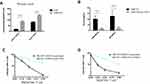
To determine the clinical relevance of LTBP4 expression in HCC, we used quantitative PCR (qPCR) to analyze the levels of LTBP4 in 20 paired HCC samples. We found lower LTBP4 expression in HCC tumor tissue compared with normal healthy tissue (Figure 2A). When we summarized the clinical information of 46 HCC patients, we found that lower LTBP4 expression was associated with a higher occurrence of metastasis events in patients (Table 2). The treatment response of 46 patients was quantified, which demonstrated that low levels of LTBP4 expression occurred in patients with partial remission or progression of HCC, which indicated that low expression of LTBP4 was linked with the progression of HCC patients (Figure 2B). In addition, we investigated the expression pattern of LTBP4 in the TCGA database and found that LTBP4 was not only expressed at low levels in HCC tumor tissue (Figure 2C), but was also associated with the overall survival rate of patients with HCC (Figure 2D).
 |
Table 2 Summary of Clinical Information of 46 HCC Patients Divided into LTBP4 Low and High Groups Based on LTBP4 Expression in mRNA Level |
Inhibition of LTBP4 Induced EMT (Epithelial–Mesenchymal Transition) Process and Secretion of Active TGFβ1
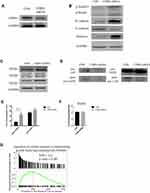
Since LTBP4 inhibition promoted cell proliferation and migration, we hypothesized that the function of LTBP4 relied on TGFβ1. As such, we designed LTBP4 shRNAs, which were transfected into Hep G2 cells (Figure 3A). After LTBP4 inhibition, the EMT marker N-cadherin, vimentin, was found to be up-regulated, while E-cadherin was down-regulated, indicating that the EMT process is activated after LTBP4 knockdown. Furthermore, the phosphorylation of genes downstream of TGFβ1, namely Smad2/3 and Smad4, were found to be increased (Figure 3B). Conversely, LTBP4 knockdown only slightly increased TGFβ1 at the protein level, which indicated that LTBP4 did not change TGFβ1 transcription (Figure 3C). In normal cells, LAP binds with TGFβ1, which is then secreted outside of the cells via a specific cutting process, through which TGFβ1 is activated.9 As such, we hypothesized that LTBP4 inhibition would lead to TGFβ1 being activated other than by transcription. We used an IP assay for LTBP4 inhibition and found that LAP binding to TGFβ1 significantly decreased (Figure 3D). Activated TGFβ1 significantly increased in this process, while total TGFβ1 was not significantly increased (Figure 3E), also the concentration of TGFβ3 is unchanged after LTBP4 is knocked down (Figure 3F). Furthermore, tumor samples from GSE14520 were divided into LTBP4 low and high expression groups, where the regulation of the TGFβ pathway was highly enriched in the LTBP4 low expression group (Figure 3G). Taken together, these results indicated that cells secreted the activated form of TGFβ1 when LTBP4 was inhibited.
Deficiency of LTBP4 Stimulated Immune Suppression Signal Through MDSC Activation
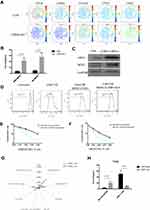
Based on previous results, we proved that LTBP4-inhibited cells could secrete more TGFβ1, which may recruit MDSCs and suppress T cell proliferation.10 As such, CyTOF was used to screen immune cell markers in LTBP4 knockout primary tumor cells. Among all the immune cell markers, CD3 and CD8A decreased while LY6G and LY6C increased in the LTBP4 knockout group (green circle in Figure 4A), suggesting that LTBP4-KO suppressed CD8+ T cells and increased PMN-MDSCs and MO-MDSCs (Figure 4B). Western blotting showed that, in LTBP4-KO primary tumors, ARG1 and NOS2 in MDSCs were all up-regulated (Figure 4C). Since both are key immunosuppressive mediators, we suspected that the MDSCs in the primary tumors were activated after LTBP4-KO. We treated the total MDSCs with conditioned medium from CON and LTBP4-KO cells, co-cultured with CD8+ T cells (MDSCs: T cells = 1:3). The conditioned medium from the CON group slightly inhibited CD8+ T cell proliferation by reducing the divided CD8+ T cells, while the conditioned medium from the LTBP4-KO group significantly inhibited CD8+ T cell proliferation (Figure 4D). The total MDSCs were then divided into PMN-MDSCs (Figure 4E) and MO-MDSCs (Figure 4F) before treating with conditioned medium from CON or LTBP4-KO cells. MO-MDSCs treated with LTBP4-KO medium showed the highest inhibition ability, indicating that LTBP4-KO induces fully functional MO-MDSCs. We also extracted hepatocellular carcinoma patients’ data from the TCGA dataset, which we divided into LTBP4 high expression and low expression groups based on the RNA-seq data. Compared with patients with high levels of LTBP4 expression, in primary tumors of patients with low LTBP4 expression, the M2 macrophages increased and the CD8+ T cells decreased (Figure 4G and H). Taken together, these results indicated that LTBP4-KO recruits MDSCs in the primary tumor and inhibits CD8+ T cell proliferation, which results in the generation of an immune suppression signal.
Activated TGFβ1 Secreted by LTBP4 Inhibition Cells Increased Fully Functioned MDSCs
To confirm whether the immune suppression signal generated by LTBP4-KO was due to the secretion of active TGFβ1 by tumor cells, we measured the total and active TGFβ1 in the primary tumors. Consistent with the in vitro results, active TGFβ1 was highly increased in the LTBP4-KO primary tumors, and total TGFβ1 also increased (Figure 5A). Then, we injected LTBP4-KO cells into SCID mice. Notably, the direct injection of the TGFβ1 receptor inhibitor decreased both PMN-MDSCs and MO-MDSCs after SB-431542 treatment (Figure 5B). We also found that the conditioned medium from LTBP4-KO cells treated with SB-431542 failed to inhibit T cell proliferation in both PMN-MDSCs (Figure 5C) and MO-MDSCs (Figure 5D).
Discussion
Due to the high rate of metastasis, in particular pulmonary metastasis, the 5-year survival rate of HCC is lower than 40%, even in patients treated surgically during the early stages of the disease. As such, the identification of genes related to metastasis is of key importance.11 Within the metastasis mechanism of liver cancer, TGFβ1 plays an important role. It not only induces EMT in tumor cells but can also activate immune suppression signals and form the pre-metastatic niche.12 Despite this, the precise role of TGFβ1 in the interaction between tumor cells and the tumor microenvironment remains unknown.
Herein, we identified a new metastasis suppressor, LTBP4, using in vitro screening techniques. The knockout of LTBP4 was found to promote Hep G2 cell migration, proliferation, and metastasis in vivo, suggesting that LTBP4 participates in liver cancer cell function. LTBP4 belongs to the LTBP family and can be further subdivided into LTBP4S, LTBP-4L1, and LTBP-4L2 at the protein level.13 LTBP is also a component of the ECM in normal cells. In the ECM, TGFβ1 binds to LAP (latency-associated peptide) and LTBP protein with no function.14 Alterations to LTBP can change the status of TGFβ1. For example, in LTBP4S KO mice, the septation of end-pulmonary balloon decreased, while the activation of TGFβ1 decreased in fibroblasts15. Our study demonstrated that, unlike in lung cells, the inhibition of LTBP4 induced EMT in tumor cells, while upregulating the expression of genes downstream of TGFβ1, namely Smad2/3 and Smad4, and changing the expression of TGFβ1 slightly. Furthermore, we proved that the inhibition of LTBP4 decreased LAP-TGFβ1 binding but increased the active form of TGFβ1.
In Alexandre Calon’s study, TGFβ1 alleviation in the tumor microenvironment stimulated cancer-associated fibroblasts (CAFs), thus generating a pre-metastatic niche in colorectal cancer.16 In our study, CyTOF was used to demonstrate that the MDSCs and CD8+ T cells changed the most in LTBP4-KO primary tumors. MDSCs are a cluster of immature cells derived from bone marrow, including immature dendritic cells, neutrophils, and macrophages.17 MDSCs can induce immune evasion via the secretion of VEGF and BEGF or the suppression of T cell proliferation.18 In our study, we demonstrated that LTBP4-KO increased MDSCs and decreased CD8+ T cells in primary tumors. At the same time, the conditioned medium from LTBP4-KO cells also inhibited the proliferation of CD8+ T cells. Notably, we found that MO-MDSCs appeared to be mainly driven by tumor cells, further inhibiting T cell proliferation. By using the TGFβ1 receptor inhibitor SB-431542, we further proved that an increase in MDSCs was caused by the secretion of TGFβ1 by tumor cells. In summary, LTBP4-KO in tumor cells activated immune suppression signals by secreting TGFβ1, highlighting LTBP4 as a potential therapeutic target.
Acknowledgments
The authors are grateful for all support from Clinical Laboratory, Beilun Branch Hospital of The First Affiliated Hospital of Medical School Zhejiang University.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Ghouri YA, Mian I, Rowe JH. Review of hepatocellular carcinoma: epidemiology, etiology, and carcinogenesis. J Carcinog. 2017;16.
2. Chen ZH, Zhang XP, Wang H, et al. Effect of microvascular invasion on the postoperative long-term prognosis of solitary small HCC: a systematic review and meta-analysis. HPB. 2019;21(8):935–944.
3. Liu Q, Li F, Zhuang Y, et al. Alteration in gut microbiota associated with hepatitis B and non-hepatitis virus related hepatocellular carcinoma. Gut Pathog. 2019;11(1):1. doi:10.1186/s13099-018-0281-6
4. Toso C, Meeberg G, Andres A, et al. Downstaging prior to liver transplantation for hepatocellular carcinoma: advisable but at the price of an increased risk of cancer recurrence–a retrospective study. Transpl Int. 2019;32(2):163–172. doi:10.1111/tri.13337
5. Yu JI, Choi GS, Do Hoon LIM, et al. Treatment of naïve HCC combined with segmental or subsegmental portal vein tumor thrombosis: liver resection versus TACE followed by radiotherapy. Anticancer Res. 2018;38(8):4919–4925. doi:10.21873/anticanres.12808
6. Li K, Chen Y, Li A, et al. Exosomes play roles in sequential processes of tumor metastasis. Int J Cancer. 2019;144(7):1486–1495. doi:10.1002/ijc.31774
7. Wu S, Zheng Q, Xing X, et al. Matrix stiffness-upregulated LOXL2 promotes fibronectin production, MMP9 and CXCL12 expression and BMDCs recruitment to assist pre-metastatic niche formation. J Exp Clin Cancer Res. 2018;37(1):99. doi:10.1186/s13046-018-0761-z
8. Nakamura K, Kassem S, Cleynen A, et al. Dysregulated IL-18 is a key driver of immunosuppression and a possible therapeutic target in the multiple myeloma microenvironment. Cancer Cell. 2018;33(4):634–648. e5. doi:10.1016/j.ccell.2018.02.007
9. Warren J, Im M, Ballesteros A, et al. Activation of latent transforming growth factor-β1, a conserved function for pregnancy-specific beta 1-glycoproteins. MHR Basic Sci Reprod Med. 2018;24(12):602–612.
10. Jayaraman P, Parikh F, Newton JM, et al. TGF-β1 programmed myeloid-derived suppressor cells (MDSC) acquire immune-stimulating and tumor killing activity capable of rejecting established tumors in combination with radiotherapy. Oncoimmunology. 2018;7(10):e1490853. doi:10.1080/2162402X.2018.1490853
11. Daher S, Massarwa M, Benson AA, et al. Current and future treatment of hepatocellular carcinoma: an updated comprehensive review. J Clin Transl Hepatol. 2018;6(1):69. doi:10.14218/JCTH.2017.00031
12. Wee I, Syn N, Sethi G, et al. Role of tumor-derived exosomes in cancer metastasis. Biochim Biophys Acta Rev Cancer. 2018.
13. Dabovic B, Robertson IB, Zilberberg L, et al. Function of latent TGFβ binding protein 4 and fibulin 5 in elastogenesis and lung development. J Cell Physiol. 2015;230(1):226–236. doi:10.1002/jcp.24704
14. Robertson IB, Rifkin DB. Regulation of the bioavailability of TGF-β and TGF-β-related proteins. Cold Spring Harb Perspect Biol. 2016;8(6):a021907. doi:10.1101/cshperspect.a021907
15. Koli K, Wempe F, Sterner-Kock A, et al. Disruption of LTBP-4 function reduces TGF-β activation and enhances BMP-4 signaling in the lung. J Cell Biol. 2004;167(1):123–133. doi:10.1083/jcb.200403067
16. Calon A, Espinet E, Palomo-Ponce S, et al. Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation. Cancer Cell. 2012;22(5):571–584. doi:10.1016/j.ccr.2012.08.013
17. Ramachandran IR, Condamine T, Lin C, et al. Bone marrow PMN-MDSCs and neutrophils are functionally similar in protection of multiple myeloma from chemotherapy. Cancer Lett. 2016;371(1):117–124. doi:10.1016/j.canlet.2015.10.040
18. Seshadri DR. Immuno-Nanotherapeutics to Inhibit Macrophage Polarization for Non-Small-Cell Lung Cancers. Case Western Reserve University; 2017.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.