Back to Journals » Journal of Asthma and Allergy » Volume 14
Disrupted Expression of Circadian Clock Genes in Patients with Bronchial Asthma
Authors Chen HC, Chen YC , Wang TN, Fang WF, Chang YC, Chen YM, Chen IY, Lin MC , Yang MY
Received 18 January 2021
Accepted for publication 19 March 2021
Published 16 April 2021 Volume 2021:14 Pages 371—380
DOI https://doi.org/10.2147/JAA.S302508
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Amrita Dosanjh
Hung-Chen Chen, 1, 2 Yung-Che Chen, 1 Tsu-Nai Wang, 3 Wen-Feng Fang, 1 Ya-Chun Chang, 1 Yu-Mu Chen, 1 I-Ya Chen, 2 Meng-Chih Lin, 1 Ming-Yu Yang 2, 4
1Division of Pulmonary and Critical Care Medicine, Department of Medicine, Chang Gung Memorial Hospital-Kaohsiung Medical Center, Chang Gung University College of Medicine, Kaohsiung, Taiwan; 2Graduate Institute of Clinical Medical Sciences, College of Medicine, Chang Gung University, Tao-Yuan, Taiwan; 3Department of Public Health, College of Health Science, Kaohsiung Medical University, Kaohsiung, Taiwan; 4Department of Otolaryngology, Kaohsiung Chang Gung Memorial Hospital, Kaohsiung, Taiwan
Correspondence: Ming-Yu Yang
Graduate Institute of Clinical Medical Sciences, College of Medicine, Chang Gung University, No. 259, Wenhua 1st Road, Guishan Dist., Taoyuan City, 33302, Taiwan
Email [email protected]
Meng-Chih Lin
Division of Pulmonary and Critical Care Medicine, Department of Medicine, Kaohsiung Chang Gung Memorial Hospital, No. 123, Dapi Road Niaosong Dist., Kaohsiung City, 83301, Taiwan
Email [email protected]
Purpose: Circadian clock is synchronized to the 24-hour day by the daily light–dark cycle and proper function of circadian rhythm is essential for many physiological processes. Disruption of circadian rhythm can affect disease processes and influence disease severity, treatment responses, and even survivorship. In this retrospective case-controlled study, we tried to explore whether expression of circadian clock genes was disturbed in patients with bronchial asthma.
Patients and Methods: We performed real-time quantitative reverse transcriptase-polymerase chain reactions to examine the expression of the nine core circadian clock genes (BMAL1, CK1ϵ, CLOCK, CRY1, CRY2, PER1, PER2, PER3, and TIM) in total leukocytes of peripheral blood collected at chest clinics from 120 patients with asthma and 60 health individuals.
Results: Expression levels of the nine circadian clock genes were significantly different between patients and healthy individuals, but not associated with the asthma control status. We also noted the difference of PER3 expression in asthmatic patients with and without nocturnal symptoms. In well-controlled asthmatics, expression of BMAL1, CK1ϵ, CLOCK, CRY1, CRY2, and PER1 was significantly lower in patients with nocturnal symptoms than those without nocturnal symptoms. However, in not well-controlled asthmatics, expression of only BMAL1, CK1ϵ, PER1, and PER2 was significantly different between patients with and without nocturnal symptoms. Binary logistic regression analysis selected BMAL1, CKIϵ, PER3, and TIM as independent factors for bronchial asthma and ROC curves showed the combined expression of these four genes enhanced the capability of predicting asthma (AUC=0.924; 95% CI=0.875– 0.958; P< 0.001).
Conclusion: Our results showed altered expression of circadian clock genes in patients with bronchial asthma and down-regulated PER3 in patients with nocturnal symptoms. Altered expression of circadian clock genes was also observed in asthmatics with or without nocturnal symptoms in well- or not well-controlled subgroups. Combined expression of BMAL1, CKIϵ, PER3, and TIM could be a potential predictor for bronchial asthma.
Keywords: bronchial asthma, nocturnal symptom, circadian clock genes, PER3
Corrigendum for this paper has been published
Introduction
Bronchial asthma is characterized by variable airflow limitation and chronic airway inflammation with a history of respiratory symptoms such as wheeze, shortness of breath, chest tightness, and cough that vary over time and in intensity.1 It is an important characteristic of asthma that the appearance of symptoms displays a circadian rhythm2 and frequently exacerbates in the early morning hour at around 4:00AM, which is also an occurrence time of sudden death.3 It has been reported that about 74% of asthmatic patients are awakened by asthma symptoms at least once per week and around 40% of patients experienced symptoms nightly.3 Nocturnal asthma is typically defined as the difference in forced expiratory volume in 1s (FEV1) and peak expiratory flow rate between daytime and night is greater than 15% in patients with asthma.4 This overnight decline of lung function causes a more severe form of the disease and is associated with airway inflammation and hyper-responsiveness.5
Circadian rhythms are intrinsic biological oscillations with an interval near 24 hours operated in mammals by the circadian clock system.6 Molecular circadian systems are found at the cellular level by an autoregulatory transcriptional-translational feedback loop operated with at least nine circadian clock genes including PER1, PER2, PER3, CLOCK, CRY1, CRY2, BMAL1, CK1ε, and TIM.7 The circadian presentations of asthma, like airflow limitation and airway hyper-responsiveness, were associated with chronic inflammation and it may be secondary to changes of the parasympathetic nervous system and endocrine system at night.8,9 Circadian clock genes also regulate inflammatory responses and contribute to lung inflammation, fibrosis, immunity, and glucocorticoid response.10−12 Therefore, increasing evidence demonstrates the molecular circadian clock is important in the pathogenesis of asthma.
The clock in the monocyte of peripheral blood mononuclear cells (PBMC) controlled the rhythmic expression of downstream genes13 and influenced on innate immunity including gating of pattern-recognition receptors response, clock-controlled cytokine response and recruitment to tissues.13–15 Rhythmic cytokine secretion from monocytes has been reported in humans under sleep/wake and laboratory conditions.16,17 In addition, a rhythm of the adaptive immune response was measured as antibody production at different times of the day.18 That hints that either T or B lymphocyte response and the adaptive immune system was associating with the molecular circadian clock.19,20 Some asthmatic patients with nocturnal symptoms had a significantly higher number of alveolar eosinophils at 4:00AM than at 4:00PM, and the increase of alveolar eosinophils correlated with a decrease in FEV1.3,21 Also, the number of macrophages, neutrophils and CD4 T-cells in the bronchoalveolar lavage fluid from patients with asthma were higher at 4:00AM than at 4:00PM, and the increase of CD4 T-cells also correlated with decreased FEV1.3,21
Recently, several studies found circadian clock underpins allergic reaction and possibly influences the periodicity of allergic disease.22–24 The induction of asthma was associated with markedly increased inflammation in the lungs in mice lacking Bmal1 expression in myeloid cells, and a higher numbers of eosinophils and increased IL-5 levels in the lung and serum.25 Since altered circadian rhythm deregulates epithelial barrier function and immune function, the two fundamental biological aspects of allergic disease, the circadian clock is strongly related to allergic disease.
For the substantial associations of circadian presentations between asthma and allergic diseases, and for the lack of reports about expression of circadian clock genes in patients with asthma, we therefore hypothesized that the expression of circadian gene expression might be disrupted in asthmatic patients and correlated with nocturnal symptoms of asthmatic patients. To test our hypothesis, we constructed a retrospective case-controlled study focusing on expression of circadian clock genes of PBMCs in patients with bronchial asthma and healthy individuals and to explore the correlation of circadian clock gene expression between difference control status and clinical presentation in these patients. In addition, we also tried to build a prediction model for asthma by finding the expression of a specific circadian clock gene in PBMCs.
Patients and Methods
Patients, Healthy Subjects, and Samples
We conducted this retrospective case-controlled study between January 2019 and December 2019 at Kaohsiung Chang Gung Memorial Hospital (K-CGMH), Taiwan. The design of this study adhered to the principles of the Declaration of Helsinki. This study was approved by the Institutional Review Board (IRB) of the CGMH Ethical Committee (IRB No. 201801642B0A3C501). Written informed consent was obtained from all participants.
Adult asthmatic patients were recruited from the Division of Pulmonary and Critical Care Medicine of K-CGMH and the Division of Chest Medicine of Kaohsiung Medical University Hospital (KMUH) in Taiwan from January 2013 to January 2015. The candidates had symptoms such as episodic breathlessness, wheezing, cough, and chest tightness and the diagnosis of asthma according to the Global Initiative for Asthma (GINA) guidelines, with or without reversible airway from spirometry that showed an increase in FEV1 of at least 12% and at least 200 mL from the prebronchodilator value. Those who met the previous criteria were regarded to have asthma if they had had at least one asthma exacerbation or needed use of asthma prescription in the 12 months before the clinics visit. Patients were excluded if they were physician-diagnosed with chronic airway obstruction, emphysema, tuberculosis, or cancer.
We obtained residual cDNA samples of PBMCs and collected clinical parameters and outcomes from 120 patients with asthma (60 men and 60 women, aged 25–73) and 60 age- and gender-matched healthy individuals (30 men and 30 women, aged 20–73). All the enrolled subjects were aged 18 years or older. Pregnant women, psychiatric patients, and individuals taking sleep pills were excluded. Subjects enrolled in this study did not experience shift work or jet lag 1 week before PB collection. The study participants were screened for eligibility and recruited from the pulmonary clinics and health examination center of K-CGMH and KMUH during the period of January 2013 through January 2015. Clinical characteristics including gender, age, smoking habits, body weight, body mass index (BMI), body fat, body height, waist, IgE, FEV1, PEF (peak expiratory flow rate), and asthma control test (ACT) were recorded. We defined the well-controlled and poor-controlled asthma as ACT scores equal to or more than 20 and below 20, respectively. Patients with nocturnal symptoms were identified in the third question of ACT, with a score lower than 5, which means asthma symptoms (wheezing, coughing, shortness of breath, chest tightness, or pain) have woken patients up at night or earlier than usual in the morning during the past 4 weeks.
Analysis of Expression of Circadian Clock Genes
The expression of the nine circadian clock genes and ACTB gene (as endogenous reference control) was analyzed using real-time quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) as previously described.26,27 The relative threshold cycle (ΔCt) of the circadian clock gene was obtained by normalizing the threshold cycle of the circadian clock gene to the ACTB gene of the same sample, ie, ΔCt = (Ct of circadian clock gene – Ct of ACTB gene). Thus, a higher ΔCt value represents a lower expression level and a higher -ΔCt value represents a higher expression level.
Statistical Analysis
We used MedCalc software (MedCalc Software Ltd, Ostend, Belgium) and Graph Pad Prism 7.04 (GraphPad, San Diego, CA, USA) for all statistical analyses. The sample size was estimated by G power software (Supplementary Figure S1). Student’s t-test was used to detect the differences between patients with bronchial asthma and healthy subjects, and Kruskal–Wallis test was used to detect the differences among participants with well- or not poor-controlled asthma and healthy subjects in each circadian clock gene expression. Post-hoc analysis was used for comparing the difference among subgroups. Mann–Whitney test was used to detect the differences between asthma patients with and without night symptoms. Receiver operating characteristic (ROC) curve was plotted and ROC area under the curve (AUC) was calculated to compare the discriminating ability of each circadian clock genes. Binary Logistic regression with multivariable analysis modelled the effects of selected independent variables on whether the expression of a specific circadian clock gene could be a predictive marker for asthma or not. The values of ΔCt or -ΔCt were used for all statistical analysis. All tests were two-sided with statistical significance set at 0.05.
Results
Characteristics of Study Participants
The characteristics, including gender, age, smoking habits, body weight, BMI, body fat, body height, and waist of the 120 patients with bronchial asthma and 60 healthy individuals are listed in Table 1. All the characteristics are not different between patients and healthy individuals. The values for IgE, FEV1, PEF, and ACT score are also listed for the patients with bronchial asthma. The characteristics of well- and not well-controlled asthmatics and asthmatics with and without night symptoms are listed in Table 2.
 |
Table 1 Characteristics of Study Participants |
 |
Table 2 Characteristics of Well- and Not Well-Controlled Asthmatics and Asthmatics with and without Night Symptoms |
Expression of Circadian Clock Genes in Patients with Bronchial Asthma
In order to explore whether expression of circadian clock genes altered in patients with bronchial asthma, we performed qRT-PCR to examine the expression of a panel of nine human circadian clock genes in the PBMCs. We found significant difference of expression of all the nine circadian clocks genes between asthmatic patients and healthy individuals (Figure 1A). Except for BMAL1, the expression of the other eight genes was down-regulated in patients with bronchial asthma. We divided the patients into with and without controlled status, but the expression of all the nine circadian clock genes was not different between these two groups of patients (Figure 1B).
Expression of Circadian Clock Genes in Asthmatic Patients with and without Night Symptoms
To investigate whether the expression of circadian clock genes correlated with night symptoms, we further divided the asthmatic patients into with and without night symptoms groups and performed a Mann–Whitney test to investigate the differential expression of circadian clock genes between these two groups. Among the nine genes, PER3 was the only gene with lower expression in the group with night symptoms (P=0.0408) (Figure 2 and Supplementary Table S1).
Expression of Circadian Clock Genes in Well- or Not Well-Controlled Asthmatics with and without Night Symptoms
We further divided the well-controlled asthmatics into groups of with and without night symptoms and found significant down-regulation of BMAL1, CK1ε, CLOCK, CRY1, CRY2, and PER1 in the group with night symptoms (Figure 3A and Supplementary Table S2). We also divided the not well-controlled asthmatics into with and without night symptoms groups, and altered expression of BMAL1, CK1ε, PER1, and PER2 was observed between these two groups (Figure 3B and Supplementary Table S2).
Predictive Ability for Bronchial Asthma Based on the Expression of Circadian Clock Genes
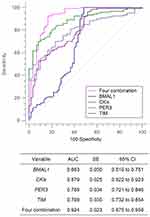
To investigate the discriminative performance of expression of circadian clock genes between the groups of asthma and healthy individuals, we constructed ROC curves and calculated the AUC for each gene. Among the nine circadian clock genes, binary logistic regression analysis selected BMAL1 (Odds ratio=0.23; 95% confidence interval (CI)=0.12–0.43; P<0.0001), CKIε (Odds ratio=12.34; 95% CI=3.92–38.77; P<0.0001), PER3 (Odds ratio=2.15; 95% CI=1.08–4.26; P=0.0293), and TIM (Odds ratio=0.45; 95% CI=0.45–0.82; P=0.0097) as independent factors for bronchial asthma (Table 3). Combined expression of BMAL1, CKIε, PER3, and TIM enhanced the prediction of bronchial asthma (Odds ratio=624.52; 95% CI=115.38–3,380.37; P<0.0001) (Table 3). We also performed ROC analysis and evaluated the AUC of ROC curves to investigate whether patients with bronchial asthma could be distinguished from healthy subjects based on their expression of circadian clock genes (Figure 4). The discriminating predictive abilities of CKIε and four combinations were found to be excellent (AUC=0.879 and 0.924, respectively), and predictive abilities of BMAL1, PER3, and TIM were acceptable (AUC range=0.683–0.798). Our results demonstrated the combined expression of BMAL1, CKIε, PER3, and TIM might have the potential to be a predictor for bronchial asthma.
 |
Table 3 Binary Logistic-Regression Analysis of Predictors of Bronchial Asthma by Expression of BMAL1, CKIε, PER3, and TIM. |
Discussion
In this case-controlled study, we found the expression of all the nine circadian clock genes was altered when compared with that in health individuals. In consistent with our finding, a recent study demonstrated significantly decreased expression of TIM in the blood of children with asthma as compared to the healthy children.28 Other than this report on the TIM gene, there have so far been no reports to directly link the deregulated circadian clock genes and bronchial asthma in humans. Obstructive sleep apnea (OSA) has been reported to be prevalent in nocturnal asthma.29 Our previous study indeed observed expressions of eight circadian clock genes (except PER1) at midnight were significantly downregulated in patients with severe OSA.30 This may imply the disruption of circadian rhythm caused altered lung function in both asthma and OSA. Deregulated Per2 and Bmal1 have been associated with inflammation or exacerbated immune function and lung functions in animal models. Recently, a study had disclosed that aberrant light/dark situation aggravated virus-induced, asthma-like inflammation.31 Bmal1-deleted mice were more susceptible to bacterial infections and inflammation,12,31,32 and developed more extensive airway inflammation than wild-type mice.33 Inflammatory factors could affect peripheral clock activity and the phase, period, and amplitude of expression of circadian clock genes in normal lung tissue were changed by exposure to cigarette smoke, viral or bacterial infection.34,35 In addition, hormonal or autonomic nerve activity synchronized peripheral clocks to the suprachiasmatic nucleus rhythm and affect the asthma activity.36 The aberrant hormone secretion or autonomic nerve activity might desynchronize peripheral clock rhythm and might affect the airway activity and allergic reaction. For example, chronic stress would be relating to arrhythmicity in Per2 expression in mice.36 Per2 gene regulates daily IL-1β production in macrophages and IFN-γ production in NK cells, altered Per2 gene thus leads to the impairment of immune function.35 The majority of asthmatic patients have an allergy reaction and several studies showed the circadian disruption can promote allergy.24,37–39 The mechanism might relate to circadian gating of epithelial barrier function and immune responses that evolved to complex environmental physical, chemical, and biological stimulants and enhance host defense after exposure of stimulants.24,40
In this study, we noted a lower expression of PER3 in asthmatic patients with night symptoms. PER3 was thought to be less important than the other two family members, PER1 and PER2, in regulating the circadian clock. However, down-regulation of PER3 has been reported in various types of human cancers,26,27,41,42 and has been associated with sleep homeostasis and mental disorders in humans. Interestingly, PER3 was also the most down-regulated gene at midnight in OSA and was selected for predicting OSA.30 Hence, our finding may add evidence to strengthen the crucial role of PER3 in fine adjustment of response in the time-keeping system in humans.
Another novel finding of this study was the combined expression of BMAL1, CKIε, PER3, and TIM has the potential to predict bronchial asthma. Although depending solely on the expression of circadian clock genes may not be sufficient to predict asthma, they did provide a new reference for evaluating asthma.
Although we have obtained novel and clinically relevant results that could give an insight into a more profound understanding of asthma, there are limitations of this study. First, all the PB samples were obtained in the daytime and only at one time point and therefore the oscillation of circadian clocks genes may not be reflected fully. Second, we did not collect cases at different treatment times to validate whether the changes of gene expression related to the disease status of patients. In addition, this is a correlation study, although we have linked circadian clock genes to with or without control and night symptoms of patients with asthma, no mechanisms of these gene functions were investigated. Due to this study is a retrospective study, we lack detailed medication, treatment response and some data, such as fractional exhaled nitric oxide (FeNO) and allergic profile. Moreover, for the retrospective study with residual samples, we did not have samples to verify our qPCR data at the protein level using either Western blot or immunohistochemical staining.
Conclusion
This is the first study that directly links circadian clock genes to human asthma. We demonstrated expression levels of the nine core circadian clock genes were altered in patients with bronchial asthma. We also discovered PER3 could be a new reference for evaluating the asthmatics patients with nocturnal symptoms and the combined expression of BMAL1, CKIε, PER3, and TIM has the potential to be a predictor for asthma. Our results may not be sufficient for clinical diagnosis of asthma presently, but they did provide new directions for future studies. Investigating the mechanisms of downregulated circadian clock genes in asthma and the molecular causal effects of circadian clock genes and asthma will also let us take a step forward to reveal the mysterious roles circadian clock genes play in asthma.
Acknowledgments
This study was funded by grants from the CGMH (Grant number: CMRPG8J0191). The authors thank the Biostatistics Center, K-CGMH for statistics work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Reddel HK, Bateman ED, Becker A, et al. A summary of the new GINA strategy: a roadmap to asthma control. Eur Respir J. 2015;46(3):622–639. doi:10.1183/13993003.00853-2015
2. Clark TJ. Diurnal rhythm of asthma. Chest. 1987;91(6 Suppl):137S–141S. doi:10.1378/chest.91.6_Supplement.137S
3. Litinski M, Scheer FA, Shea SA. Influence of the circadian system on disease severity. Sleep Med Clin. 2009;4(2):143–163. doi:10.1016/j.jsmc.2009.02.005
4. Martin RJ. Location of airway inflammation in asthma and the relationship to circadian change in lung function. Chronobiol Int. 1999;16(5):623–630. doi:10.3109/07420529908998731
5. Sutherland ER. Nocturnal asthma. J Allergy Clin Immunol. 2005;116(6):
6. Albrecht U. Timing to perfection: the biology of central and peripheral circadian clocks. Neuron. 2012;74(2):246–260. doi:10.1016/j.neuron.2012.04.006
7. Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci. 2012;35:445–462. doi:10.1146/annurev-neuro-060909-153128
8. Greenberg H, Cohen RI. Nocturnal asthma. Curr Opin Pulm Med. 2012;18(1):57–62. doi:10.1097/MCP.0b013e32834d098e
9. Sundar IK, Yao H, Sellix MT, Rahman I. Circadian molecular clock in lung pathophysiology. Am J Physiol Lung Cell Mol Physiol. 2015;309(10):L1056–L1075. doi:10.1152/ajplung.00152.2015
10. Silver AC, Arjona A, Walker WE, Fikrig E. The circadian clock controls toll-like receptor 9-mediated innate and adaptive immunity. Immunity. 2012;36(2):251–261. doi:10.1016/j.immuni.2011.12.017
11. Gibbs JE, Beesley S, Plumb J, et al. Circadian timing in the lung; a specific role for bronchiolar epithelial cells. Endocrinology. 2009;150(1):268–276. doi:10.1210/en.2008-0638
12. Pekovic-Vaughan V, Gibbs J, Yoshitane H, et al. The circadian clock regulates rhythmic activation of the NRF2/glutathione-mediated antioxidant defense pathway to modulate pulmonary fibrosis. Genes Dev. 2014;28(6):548–560. doi:10.1101/gad.237081.113
13. Keller M, Mazuch J, Abraham U, et al. A circadian clock in macrophages controls inflammatory immune responses. Proc Natl Acad Sci USA. 2009;106(50):21407–21412. doi:10.1073/pnas.0906361106
14. Gibbs JE, Blaikley J, Beesley S, et al. The nuclear receptor REV-ERBα mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc Natl Acad Sci USA. 2012;109(2):582–587. doi:10.1073/pnas.1106750109
15. Hayashi M, Shimba S, Tezuka M. Characterization of the molecular clock in mouse peritoneal macrophages. Biol Pharm Bull. 2007;30(4):621–626. doi:10.1248/bpb.30.621
16. Born J, Lange T, Hansen K, Molle M, Fehm HL. Effects of sleep and circadian rhythm on human circulating immune cells. J Immunol. 1997;158(9):4454–4464.
17. Rahman SA, Castanon-Cervantes O, Scheer FA, et al. Endogenous circadian regulation of pro-inflammatory cytokines and chemokines in the presence of bacterial lipopolysaccharide in humans. Brain Behav Immun. 2015;47:4–13. doi:10.1016/j.bbi.2014.11.003
18. Fernandes G, Halberg F, Yunis EJ, Good RA. Circadian rhythmic plaque-forming cell response of spleens from mice immunized with SRBC. J Immunol. 1976;117(3):962–966.
19. Bollinger T, Leutz A, Leliavski A, et al. Circadian clocks in mouse and human CD4+ T cells. PLoS One. 2011;6(12):e29801. doi:10.1371/journal.pone.0029801
20. Silver AC, Arjona A, Hughes ME, Nitabach MN, Fikrig E. Circadian expression of clock genes in mouse macrophages, dendritic cells, and B cells. Brain Behav Immun. 2012;26(3):407–413. doi:10.1016/j.bbi.2011.10.001
21. Kelly EA, Houtman JJ, Jarjour NN. Inflammatory changes associated with circadian variation in pulmonary function in subjects with mild asthma. Clin Exp Allergy. 2004;34(2):227–233. doi:10.1111/j.1365-2222.2004.01866.x
22. Christ P, Sowa AS, Froy O, Lorentz A. The circadian clock drives mast cell functions in allergic reactions. Front Immunol. 2018;9:1526. doi:10.3389/fimmu.2018.01526
23. Nakao A. Clockwork allergy: how the circadian clock underpins allergic reactions. J Allergy Clin Immunol. 2018;142(4):1021–1031. doi:10.1016/j.jaci.2018.08.007
24. Nakao A. Circadian regulation of the biology of allergic disease: clock disruption can promote allergy. Front Immunol. 2020;11:1237. doi:10.3389/fimmu.2020.01237
25. Zasłona Z, Case S, Early JO, et al. The circadian protein BMAL1 in myeloid cells is a negative regulator of allergic asthma. Am J Physiol Lung Cell Mol Physiol. 2017;312(6):L855–860. doi:10.1152/ajplung.00072.2017
26. Yang MY, Chang JG, Lin PM, et al. Downregulation of circadian clock genes in chronic myeloid leukemia: alternative methylation pattern of hPER3. Cancer Sci. 2006;97(12):1298–1307. doi:10.1111/j.1349-7006.2006.00331.x
27. Yang MY, Yang WC, Lin PM, et al. Altered expression of circadian clock genes in human chronic myeloid leukemia. J Biol Rhythms. 2011;26(2):136–148. doi:10.1177/0748730410395527
28. Langwinski W, Sobkowiak P, Narozna B, et al. Association of circadian clock TIMELESS variants and expression with asthma risk in children. Clin Respir J. 2020;14(12):1191–1200. doi:10.1111/crj.13260
29. Shigemitsu H, Afshar K. Nocturnal asthma. Curr Opin Pulm Med. 2007;13(1):49–55. doi:10.1097/MCP.0b013e328010a890
30. Yang MY, Lin PW, Lin HC, et al. Alternations of circadian clock genes expression and oscillation in obstructive sleep apnea. J Clin Med. 2019;8(10):1634. doi:10.3390/jcm8101634
31. Ehlers A, Xie W, Agapov E, et al. BMAL1 links the circadian clock to viral airway pathology and asthma phenotypes. BMAL1 links the circadian clock to viral airway pathology and asthma phenotypes. Mucosal Immunol. 2018;11(1):97–111. doi:10.1038/mi.2017.24
32. Nguyen KD, Fentress SJ, Qiu Y, Yun K, Cox JS, Chawla A. Circadian gene Bmal1 regulates diurnal oscillations of Ly6C(hi) inflammatory monocytes. Science. 2013;341(6153):1483–1488. doi:10.1126/science.1240636
33. Gibbs J, Ince L, Matthews L, et al. An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action. Nat Med. 2014;20(8):919–926. doi:10.1038/nm.3599
34. Hwang JW, Sundar IK, Yao H, Sellix MT, Rahman I. Circadian clock function is disrupted by environmental tobacco/cigarette smoke, leading to lung inflammation and injury via a SIRT1-BMAL1 pathway. FASEB J. 2014;28(1):176–194. doi:10.1096/fj.13-232629
35. Annamneedi VP, Park JW, Lee GS, Kang TJ. Cell autonomous circadian systems and their relation to inflammation. Biomol Ther (Seoul). 2021;29(1):31–40. doi:10.4062/biomolther.2020.215
36. Nakamura Y, Ishimaru K, Tahara Y, Shibata S, Nakao A. Disruption of the suprachiasmatic nucleus blunts a time of day-dependent variation in systemic anaphylactic reaction in mice. J Immunol Res. 2014;2014:474217. doi:10.1155/2014/474217
37. Kawauchi T, Ishimaru K, Nakamura Y, et al. Clock-dependent temporal regulation of IL-33/ST2-mediated mast cell response. Allergol Int. 2017;66(3):472–478. doi:10.1016/j.alit.2017.02.004
38. Nakamura Y, Nakano N, Ishimaru K, et al. Circadian regulation of allergic reactions by the mast cell clock in mice. J Allergy Clin Immunol. 2014;133(2):568–575. doi:10.1016/j.jaci.2013.07.040
39. Ihara T, Mitsui T, Nakamura Y, et al. Clock genes regulate the circadian expression of Piezo1, TRPV4, Connexin26, and VNUT in an ex vivo mouse bladder mucosa. PLoS One. 2017;12(1):e0168234. doi:10.1371/journal.pone.0168234
40. Curtis AM, Bellet MM, Sassone-Corsi P, O’Neill LA. Circadian clock proteins and immunity. Immunity. 2014;40(2):178–186. doi:10.1016/j.immuni.2014.02.002
41. Yang MY, Lin PM, Hsiao HH, et al. Up-regulation of PER3 expression is correlated with better clinical outcome in acute leukemia. Anticancer Res. 2015;35(12):6615–6622.
42. Hsu CM, Lin SF, Lu CT, Lin PM, Yang MY. Altered expression of circadian clock genes in head and neck squamous cell carcinoma. Tumour Biol. 2012;33(1):149–155.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.