Back to Journals » Clinical, Cosmetic and Investigational Dermatology » Volume 16
Discussion on the Antipruritic Mechanism of Qiwei Antipruritic Based on Network Pharmacology and Molecular Docking Technology
Authors Wang L, Deng T, Liu Y, Cheng H
Received 14 September 2023
Accepted for publication 24 October 2023
Published 15 November 2023 Volume 2023:16 Pages 3295—3307
DOI https://doi.org/10.2147/CCID.S435800
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Jeffrey Weinberg
Luoxi Wang,1 Tinghan Deng,1 Ying Liu,1 Hongbin Cheng2
1Clinical Research on Skin Diseases School of Clinical Medicine, Chengdu University of TCM, Chengdu, People’s Republic of China; 2Dermatology of Department, Hospital of Chengdu University of Traditional Chinese Medicine, Chengdu, People’s Republic of China
Correspondence: Hongbin Cheng, Email [email protected]
Objective: To explore the mechanism of Qiwei antipruritic by using network pharmacology and molecular docking technology.
Methods: The components and related targets of Qiwei antipruritic were screened by using the traditional Chinese medicine system pharmacology database (TCMSP and symmap databases). GeneCards and OMIM databases were used to screen itch-related targets. The protein–protein interaction (PPI) network between active ingredient targets and pruritus disease targets was constructed using STRING database. Cytoscape 3.8.0 software was used to draw the visualization network of “drug-component-target-signaling pathway” and screen the core targets. Gene ontology (GO) functional annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed using R software. AutoDock vina software was used to perform molecular docking of key targets and their corresponding key components.
Results: There were 44 main components of Qiwei antipruritic compound, 118 corresponding targets and 3869 itch-related genes. A total of 82 predicted targets of Qiwei antipruritic in the treatment of pruritus were obtained. Eleven key targets were screened. Among the 23 KEGG enriched pathways, 12 signaling pathways were related to skin pruritus. Molecular docking results showed that the core components of Qiwei antipruritic, including quercetin, kaempferol, β-sitosterol, stigmasterol, luteolin, and preskimmianine, had good binding ability with ESR1, PPARG, IL6, TP53, and EGFR, and the docking scores were all less than − 4.
Conclusion: The mechanism of Qiwei antipruritic may be related to histamine activation mechanism, calcium channel mechanism, inhibition of inflammatory signaling pathway, inhibition of neurotransmitters, and regulation of immune pathways. The traditional Chinese medicine compound Qiwei antipruritic can treat clinical pruritus through multiple targets and pathways.
Keywords: Qiwei antipruritic, traditional Chinese medicine compound, network pharmacology, molecular docking, mechanism of action
Introduction
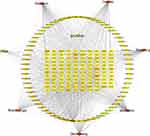
Pruritus (also known as scratching or pruritus) is a very common symptom seen in a variety of skin and systemic diseases. Atopic dermatitis, psoriasis, contact dermatitis, urticaria, lichen vulgaris, mycosis fungoides, scars, autoimmune diseases, and kidney or liver diseases may all produce clinical manifestations of pruritus, which seriously affect the quality of life of patients.1 With the continuous research on the mechanism of pruritus, new progress has been made in the understanding and treatment of pruritus, but the treatment of skin pruritus still faces great challenges. At present, antihistamines or steroids are still the main first-line drugs in clinical practice.2,3 However, most of them are single targets. Antihistamines are often not effective for chronic pruritus or non-histamine-dependent pruritus, with side effects such as drowsiness, dry mouth, dry eyes, and urinary retention.4,5 While long-term and excessive use of topical steroids may lead to skin thinning, rupture, skin pigmentation, acne, and easy bleeding.6 With abundant resources and diverse structure of active ingredients, traditional Chinese medicine (TCM) has a broad prospect in the treatment of skin pruritus. Network pharmacology is an emerging bioinformatics method for studying target molecules in recent years. It was first proposed by Andrew L Hopkins.7 In this paper, the network pharmacology research method and molecular docking were used to analyze the potential targets and mechanism of modified Qiwei antipruritic in the treatment of skin pruritus, which is of great significance for further research on the mechanism of antipruritic traditional Chinese medicine and the study of its active ingredients for skin pruritus. The “Qiwei antipruritic” formula is made up of seven ingredients: Cnidii Fructus, Kochiae Fructus, Dictamni Cortex, Portulacae Herba, Senecionis Scandentis Hebra, Vespae Nidus, and Zanthoxyli Pericarpium. Among them, Cnidii Fructus (She Chuang zi) is known for its properties to dry dampness, eliminate parasites, and relieve itching. Kochiae Fructus (Di fu zi) is associated with the kidney and bladder pathways, and it helps to clear heat, reduce inflammation, and remove damp heat from the lower part of the body. These two, combined with Dictamni Cortex (Bai Xian pi) and Portulacae Herba (Ma Chi xian), enhance the effects of removing dampness and itchiness. Dictamni Cortex (Bai Xian pi) helps clear heat and dry dampness, while Portulacae Herba (Ma Chi xian) and Senecionis Scandentis Hebra (Qian Li guang) work together to clear heat and detoxify, focusing on expelling damp-related toxins. Vespae Nidus (Feng fang) and Zanthoxyli Pericarpium (Hua jiao) target toxins, kill parasites, and alleviate itching. Overall, this formula is effective in drying dampness, detoxifying, clearing heat, and relieving itching, and it has shown great clinical results. At present, the mechanism of action of this drug composition on skin pruritus remains to be investigated. Several studies have shown that network pharmacology approaches have been successfully applied to the mechanistic study of many traditional Chinese medicines (Figure 1).8,9
 |
Figure 1 Framework based on an integration strategy of network pharmacology. |
Materials and Methods
Collection and Screening of Main Chemical Components of Qiwei Antipruritic
From the Traditional Chinese Medicine System Pharmacology Database and Analysis Platform (TCMSP, https://old.tcmsp-e.com/tcmsp.php), Traditional Chinese Medicine Integrated Pharmacology Research Platform (TCMIP, http://www.tcmip.cn/TCMIP/index.php/Home/), symptom map database (http://symmap.org/search/) to retrieve the collection 7 itching of main chemical composition. The oral bioavailability (OB) ≥30% and drug-like properties (DL) ≥0.18 were used as screening conditions to further screen the main chemical components of the collected Qiwei antipruritic to obtain the active ingredients.10
Collection of Active Ingredient Targets of Qiwei Antipruritic and Skin Pruritus Gene Targets
The active ingredients in Qiwei antipruritic were collected and sorted out in TCMSP database, and the targets of the active ingredients were obtained. Using “PRURITUS” as the key word, it was found in the Human Gene Database (gene-cards)11 and the online Mendelian Inheritance in Man database (OMIM). (www.Omim.org/)12 to search for itch-related genes.
Construction of Active Ingredient-Disease Target-Signaling Pathway Network of Qiwei Antipruritic
The active ingredients and corresponding targets contained in the previous traditional Chinese medicine compound were collected and sorted out, and the global protein resource database (Uni Prot, https://www.uni-prot.org/) was used to standardize the protein target data.13 Then Cytoscape 3.8.0 software was used to construct the visual pathway network of “drug pair-active ingredients-target-signaling pathway”, and the degree values between compounds and targets were analyzed.14
Construction of Protein–Protein Interaction Network (PPI) and Screening of Core Targets
The collected targets of Chinese medicine active ingredients and itch disease target genes were imported into the STRING database (https://cn.string-db.org/), and the species was set as “Homo sapiens” with confidence interval≥0.99, the PPI network was constructed.15 The classical Degree value was used as the standard to measure the importance of nodes in the PPI network, and the Cy-to NCA plug-in in Cytoscape software was used to calculate the degree value of the above nodes, and the core nodes were screened by the degree value.
GO Biological Analysis and KEGG Pathway Enrichment Analysis Were Performed
The key targets predicted in the previous step were analyzed using GOcluster package in R software for gene ontology (GO) functional annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG, https://www.kegg.jp/) pathway enrichment analysis.16 The species was set as “Homo sapiens”, followed by P<0.05 was selected as the conditional screening, and the P value was ranked from small to large. The top 10 items of biological process (BP), cellular component (CC), and molecular function (MF) in GO functional annotation and the top 23 pathways in KEGG pathway enrichment were selected for visual analysis. Visual drawing display is implemented through the GOplot package.17 The aim of this study is to clarify the role of the target proteins of the intersection between Qiwei antipruritic and pruritus in gene function and signal transduction pathways.
Molecular DOCKING Validation
The key targets with the largest number of enrichment pathways were docked with the key components of traditional Chinese medicine compound. The PDB structure of key target proteins was downloaded from the PDB database, and the structure of PDB proteins and key components was preprocessed using AutoDock Vina software. The key components after pretreatment were docked with the lattice file generated by PDB protein pretreatment to verify the binding activity of key targets and key components, and the docking scores were obtained.18
Results
Screening of Active Ingredients of Qiwei Antipruritic
After collecting from the TCMSP and TCMIP databases, the active ingredients of Portulacae Herba, Zanthoxyli Pericarpium, Dictamni Cortex, Kochiae Fructus, and Cnidii Fructus are 54, 101, 633, 19, and 114, respectively. Senecionis Scandentis Hebra and Vespae Nidus have 29 and 47 active ingredients, respectively, after collecting from the SymMap database. Based on the criteria of OB≥30% and DL≥0.18, the number of active ingredients screened are 10 for Portulacae HerbaPortulaca oleracea, 5 for Zanthoxyli PericarpiumZanthoxylum bungeanum 18 for Dictamni CortexPaeonia lactiflora, 2 for Kochiae FructusGeum aleppicum, 19 for Cnidii FructusCnidium monnieri, 12 for Senecionis Scandentis HebraSenecio scandens, and 5 for Vespae Nidus After compiling and eliminating duplicate ingredients and checking in the PubChem database, a total of 44 effective active ingredients were obtained. See Table 1.
 |
Table 1 Table of Active Ingredients of Qiwei Antipruritic |
Collection of Component Targets of Qiwei Antipruritic and Itch Related Gene Targets
The corresponding targets of active ingredients of Qiwei antipruritic were collected through the TCMSP database, and 118 targets were obtained after removing duplicate data. A total of 3869 itch-related disease targets were collected from GeneCards and OMIM databases. Using Venn diagram online tool, a total of 82 disease-related and drug-related targets were screened, as shown in Figure 2.
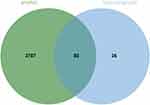 |
Figure 2 Venn diagram of disease targets and drug targets. |
Construction of Active Ingredient-Disease Target-Signaling Pathway Network of Qiwei Antipruritic
The active components of drugs, targets, and itch-related gene targets collected in the previous two steps were used to construct a visual pathway network diagram of “drug components-target-signaling pathway” using Cytoscape 3.8.0 software, and the Degree values between compounds and targets were analyzed, as shown in Figure 3.
Construction of “Disease-Drug” Protein Interaction Network and Screening of Core Targets
The predicted targets were uploaded to the string 11.0 online database (https:string-db.org/) to obtain the corresponding PPI information. The node degree value was calculated by CytoNCA plug-in in Cytoscape 3.8.0. If the node degree value was more than 2 times of the median of all nodes in the network, it could be used as a core node. The median degree value of all nodes in this study was 16, thus identifying 11 key targets in the drug-disease intersection interaction targets: IL6, VEGFA, IL1B, TP53, EGFR, PTGS2, ESR1, EGF, PPARG, CCL2, MMP2, see Figure 4. According to the degree, the targets of Qiwei antipruritic in the treatment of pruritus with D value more than twice the median were listed, see Table 2.
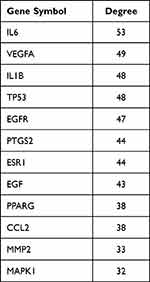 |
Table 2 Key PPI Network Targets of Qiwei Antipruritic in the Treatment of Pruritus |
 |
Figure 4 PPI network of Qiwei antipruritic in the treatment of pruritus. |
Results of GO and KEGG Enrichment Analysis
GO analysis showed that biological processes (BP) included response to nutrient levels, response to drugs, reactive oxide metabolism, response to steroid hormones, second messenger mediated signal transduction, response to purine compounds, response to lipopolysaccharide, and response to estradiol. Cellular components (CC) include membrane sheaths, membrane microdomains, membrane regions, postsynaptic membranes, caves, intrinsic components of postsynaptic membranes, synaptic membranes, etc. Molecular functions (MF) include G-protein-coupled amine receptor activity, nuclear receptor activity, ligand-activated transcription factor activity, G-protein-coupled neurotransmitter receptor activity, steroid hormone receptor activity, acetylcholine receptor activity, postsynaptic neurotransmitter receptor activity, steroid binding, serine hydrolase activity, and serine endonuclease activity. See Figure 5.
 |
Figure 5 GO analysis results. Notes: Orange bars represent biological processes (BP), green bars represent cellular components (CC), and blue bars represent molecular functions (MF). |
KEGG analysis showed that APK1, MAPK14, IL1B, IL6, MAPK8, EGFR, VEGFA, and other targets were enriched more frequently. The main enriched pathways were pathways of neurodegeneration-multiple diseases, neuroactive ligand–receptor interaction and PI3K-Akt signaling pathway, calcium signaling pathway, AGE-RAGE signaling pathway in diabetic complications, TNF signaling pathway, IL-17 signaling pathway, Th17 cell differentiation and other 23 pathways. See Figure 6. The above 12 signaling pathways are related to skin pruritus, which indicates that the composition of seven antipruritic drugs has the potential to treat skin pruritus. Skin pruritus is a common clinical symptom. The process of its production is complex and the pathways are diverse, which is related to a variety of internal and external factors, including chemicals, neurotransmitters, and inflammatory mediators.
 |
Figure 6 KEGG analysis results. |
Docking Results of Key Components of Qiwei Antipruritic and Itch-Related Key Target Proteins
We selected six core targets (EGF, EGFR, ESR1, IL6, PPARG, and TP53) and six component small molecules with high activity (beta-sitosterol, kaempferol, luteolin, preskimmianine, quercetin, and Stigmasterol) Through the molecular docking heat map, a total of 28 groups of proteins with docking scores lower than −5 were found, and the best docking score was EGFR, which proved that the active ingredients in Qiwei antipruritic had good docking activity with the key targets causing pruritus. See Figure 7.
 |
Figure 7 Heat map of docking scores. |
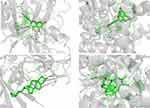
We selected four groups of typical molecular docking diagrams for display. See Figure 8. Figure 8A shows the docking of EGFR and quercetin small molecule, which can be seen to form hydrogen bonds with MET, THR, and GLU on the protein. Figure 8B shows the docking of ESR1 and luteolin small molecule. It can be seen that the small molecule forms hydrogen bonds with ARG and GLU on the protein. Figure 8C shows IL6 docking with Stigmasterol small molecule, which can be seen that the small molecule forms hydrogen bonds with ASP on the protein. Figure 8D shows PPARG docking with Stigmasterol small molecule, which can be seen that the small molecule forms hydrogen bonds with VAL on the protein.
Discussion
Through bioinformatics methods, the potential mechanism of Qiwei antipruritic in the treatment of pruritus was systematically studied. We detected 44 active ingredients and predicted 118 targets in the seven herbs of Qiwei antipruritic. Network analysis showed that Qiwei antipruritic affected pruritus by regulating IL6, VEGFA, IL1B, TP53, EGFR, PTGS2, ESR1, EGF, PPARG, CCL2, MMP2, and other targets. GO and KEGG enrichment analysis showed that the antipruritic mechanism of Qiwei antipruritic may be related to histamine activation mechanism, calcium channel mechanism, anti-inflammation, inhibition of neurotransmitters, and regulation of immune pathways. The molecular docking results of key components and core targets showed that Qiwei antipruritic had high activity.
It should be noted that EGFR had higher scores and better activity in docking with the component small molecules of Qiwei antipruritic, so we can assume that the main components of Qiwei antipruritic produce antipruritic effects mainly through EGFR. The role of EGFR in pruritus is mainly involved in two aspects: 1. Keratinization and skin barrier function: It has now been shown that both EGF expression and subsequent EGFR phosphorylation are significantly increased, which leads to tyrosine phosphorylation of JAK2, which in turn leads to phosphorylation of STAT1 and STAT5, and subsequent cell migration and proliferation.19 Although there is currently no direct evidence that deficiency or dysfunction of EGFR may contribute to skin pruritus, the keratinization and skin barrier function of EGFR are closely associated with pruritus production. For example, certain EGFR inhibitors, a class of drugs used to treat cancer, are known to trigger skin side effects, including rash and pruritus.20 Skin infections in cancer patients treated with EGFR inhibitors have also been reported.21 2. Regulation of inflammation and immune response: EGFR is also involved in the regulation of inflammation and immune response. When activated, the EGFR signaling pathway can induce the production of various inflammatory mediators, such as cytokines and chemokines, which can further stimulate nerve endings in the skin and produce pruritus. Some scholars have studied the role of EGFR ligand, Epiregulin, in the susceptibility of pulmonary tuberculosis, and also discussed its role in the regulation of inflammation and immune response, which is related to skin pruritus.22 There are also articles investigating the role of EGFR in the inflammatory response of the skin, including the induction of chemokine expression in keratinocytes, which may be associated with pruritus.23
Therefore, we can assume that the mechanism of Qiwei antipruritic is mainly through the repair of cutin and skin barrier and the regulation of inflammation and immune response, but it is not excluded that some of the active ingredients may also act through the mechanism of histamine activation, calcium channel mechanism, and the inhibition of neurotransmitters.
Conclusion
In summary, this study used network pharmacology to analyze the effective targets and action pathways of Qiwei antipruritic, and preliminarily explored the target action pathway and molecular mechanism of Qiwei antipruritic in the treatment of pruritus. The main active ingredients are quercetin, kaempferol, β-sitosterol, stigmasterol, luteolin, and pralocine, etc. The key targets are IL6, VEGFA, IL1B, TP53, EGFR, PTGS2, ESR1, EGF, PPARG, etc. Through GO and KEGG enrichment analysis, it was found that there were 12 pathways related to skin pruritus. Through molecular docking, it was found that the main active ingredients of Qiwei antipruritic had a high docking score with key targets related to pruritus, which confirmed the mechanism of action of Qiwei antipruritic with good antipruritic effect in clinical practice. The main mechanism of its action may be to repair the cuticle and skin barrier, regulate inflammation and immune response mechanisms, and is related to histamine activation mechanism, calcium channel mechanism, and inhibition of neurotransmitters. This reconfirms the effectiveness of multi-target and multi-pathway therapy of TCM. Due to the data mining and research based on network database, there are many influencing factors, which need to be verified by experiments in the next step, which will open up a new way for the treatment of pruritus diseases.
Data Sharing Statement
All data are available upon request to the corresponding author.
Acknowledgments
This work was supported by grants from the “Hundred Talents Project” of the Aliated Hospital of Chengdu University of Traditional Chinese Medicine, No. P2020042. A randomized controlled clinical study on the treatment of diabetes-associated skin pruritus with Qiwei antipruritic San and its mechanism, No. 2022YFS0413. Clinical evaluation of Picosure 755 honeycomb picosecond laser combined with Qilu Meibai ointment in the treatment of chloasma and experimental study on melanocyte metabolism and tyrosinase expression in mouse model, No. 2023YFS0331.
Disclosure
The authors report no conflicts of interest in this work. This study was approved by the Medical Ethics Committee of the Affiliated Hospital of Chengdu University of Traditional Chinese Medicine.
References
1. Elke W, Szepietowski Jacek C, Dalgard Florence J, et al. European S2k guideline on chronic pruritus. Acta Derm Venereol. 2019;99:469–506.
2. Robert S, Davis dawn M, Cohen David E, et al. Guidelines of care for the management of atopic dermatitis: section 3. Management and treatment with phototherapy and systemic agents. J Am Acad Dermatol. 2014;71(2):327–349. doi:10.1016/j.jaad.2014.03.030
3. Simons FER. Advances in H 1 -Antihistamines. N Engl J Med. 2004;351(21):2203–2217. doi:10.1056/NEJMra033121
4. K CM, Maurer M, R SFE, et al. Risk of first-generation H 1 -antihistamines: a GA 2 LEN position paper. Allergy. 2010;65(4):459–466. doi:10.1111/j.1398-9995.2009.02325.x
5. Simon FER, Simons KJ. H1 antihistamines: current status and future directions. World Allergy Organ J. 2008;1(9):145–155. doi:10.1186/1939-4551-1-9-145
6. Hengge Ulrich R, Thomas R, Schwartz Robert A, et al. Adverse effects of topical glucocorticosteroids. J Am Acad Dermatol. 2006;54(1):1–15; quiz 16–8. doi:10.1016/j.jaad.2005.01.010
7. Hopkins AL. Network pharmacology. Nat Biotechnol. 2007;25(10):1110–1111. doi:10.1038/nbt1007-1110
8. Wu Z, Yang T, Ma H. Molecular mechanism of modified Huanglian Wendan decoction in the treatment of polycystic ovary syndrome. Medicine (Baltimore). 2023;102(15):e33212. doi:10.1097/MD.0000000000033212
9. Yang J, Tian S, Zhao J, et al. Exploring the mechanism of TCM formulae in the treatment of different types of coronary heart disease by network pharmacology and machining learning. Pharmacol Res. 2020;159:105034. doi:10.1016/j.phrs.2020.105034
10. A LC, Lombardo F, W DB, et al. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2001;46:3–26.
11. Marilyn S, Irina D, Justin A, et al. GeneCards Version 3: the human gene integrator. Database (Oxford). 2010;2010:baq020.
12. Taila H, Tuğçe B B, Rojas Samantha K, et al. The unsolved rare genetic disease atlas? An analysis of the unexplained phenotypic descriptions in OMIM®. Am J Med Genet C Semin Med Genet. 2018;178:458–463.
13. Dimmer Emily C, Huntley Rachael P, Yasmin A-F, et al. The UniProt-GO annotation database in 2011. Nucleic Acids Res. 2012;40(D1):D565–70. doi:10.1093/nar/gkr1048
14. Jeong H, P MS, L BA, et al. Lethality and centrality in protein networks. Nature. 2001;411(6833):41–42. doi:10.1038/35075138
15. Tang Y, Min L, Jianxin W, et al. CytoNCA: a cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems. 2015;127:67–72. doi:10.1016/j.biosystems.2014.11.005
16. Guangchuang Y, Li-Gen W, Yanyan H, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–7.
17. Wencke W, Fátima S, Mercedes R. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics. 2015;31(17):2912–4.
18. Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi:10.1002/jcc.21334
19. Jere SW, Houreld NN, Abrahamse H. Photobiomodulation at 660nm stimulates proliferation and migration of diabetic wounded cells via the expression of epidermal growth factor and the JAK/STAT pathway. J Photochem Photobiol B. 2018;179:74–83. doi:10.1016/j.jphotobiol.2017.12.026
20. Clabbers JMK, Boers–Doets CB, Gelderblom H, et al. Xerosis and pruritus as major EGFRI-associated adverse events. Support Care Cancer. 2016;24(2):513–521. doi:10.1007/s00520-015-2781-y
21. Eilers ER, Gandhi M, Patel JD, et al. Dermatologic infections in cancer patients treated with epidermal growth factor receptor inhibitor therapy. J Natl Cancer Inst. 2010;102(1):47–53. doi:10.1093/jnci/djp439
22. White MJ, Tacconelli A, Chen JS, et al. Epiregulin (EREG) and human V-ATPase (TCIRG1): genetic variation, ethnicity and pulmonary tuberculosis susceptibility in Guinea-Bissau and The Gambia. Genes Immun. 2014;15(6):370–377. doi:10.1038/gene.2014.28
23. Francesca M, Valentina M, Giampiero G, et al. Blockade of the EGF receptor induces a deranged chemokine expression in keratinocytes leading to enhanced skin inflammation. Am J Pathol. 2003;163(1):303–312. doi:10.1016/S0002-9440(10)63654-1
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.