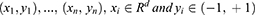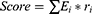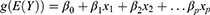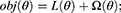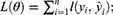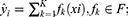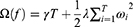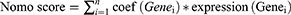Back to Journals » Clinical, Cosmetic and Investigational Dermatology » Volume 17
Discovering and Validating Cuproptosis-Associated Marker Genes for Accurate Keloid Diagnosis Through Multiple Machine Learning Models
Authors Guo Z, Yu Q, Huang W, Huang F, Chen X, Wei C
Received 8 October 2023
Accepted for publication 22 January 2024
Published 31 January 2024 Volume 2024:17 Pages 287—300
DOI https://doi.org/10.2147/CCID.S440231
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Jeffrey Weinberg
Zicheng Guo,1,2,* Qingli Yu,1,* Wencheng Huang,1 Fengyu Huang,1 Xiurong Chen,1 Chuzhong Wei1,2
1Department of Orthopaedics, Huizhou First Hospital, Huizhou, People’s Republic of China; 2Department of Orthopaedics, Southern Medical University, Guangzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Chuzhong Wei, Department of Orthopaedics, Southern Medical University, Guangzhou, People’s Republic of China, Email [email protected]
Background: Keloid is a common condition characterized by abnormal scarring of the skin, affecting a significant number of individuals worldwide.
Objective: The occurrence of keloids may be related to the reduction of cell death. Recently, a new cell death mode that relies on copper ions has been discovered. This study aimed to identify novel cuproptosis-related genes that are associated with keloid diagnosis.
Methods: We utilized several gene expression datasets, including GSE44270 and GSE145725 as the training group, and GSE7890, GSE92566, and GSE121618 as the testing group. We integrated machine learning models (SVM, RF, GLM, and XGB) to identify 10 cuproptosis-related genes (CRGs) for keloid diagnosis in the training group. The diagnostic capability of the identified CRGs was validated using independent datasets, RT-qPCR, Western blotting, and IHC analysis.
Results: Our study successfully categorized keloid samples into two clusters based on the expression of cuproptosis-related genes. Utilizing WGCNA analysis, we identified 110 candidate genes associated with cuproptosis. Subsequent functional enrichment analysis results revealed that these genes may play a regulatory role in cell growth within keloid tissue through the MAPK pathway. By integrating machine learning models, we identified CRGs that can be used for diagnosing keloid. The diagnostic efficacy of CRGs was confirmed using independent datasets, RT-qPCR, Western blotting, and IHC analysis. GSVA analysis indicated that high expression of CRGs influenced the gene set related to ECM receptor interaction.
Conclusion: This study identified 10 cuproptosis-related genes that provide insights into the molecular mechanisms underlying keloid development and may have implications for the development of targeted therapies.
Keywords: machine learning, cuproptosis, keloid, novel biomarker
Graphical Abstract:
Introduction
Keloid is a skin condition characterized by the proliferation of fibroblasts and the accumulation of extracellular matrix (ECM), resulting in significant impact on skin appearance and psychological well-being.1 Fibroblasts play a key role in the development of keloids, contributing to chronic inflammation and excessive ECM deposition.2,3 Various fibrotic growth factors, including FGF, TGFβ, VEGF, and PDGF, are known to drive fibroblast migration, invasion, and ECM deposition.4,5 Keloid disease has a significant global impact, affecting millions of individuals.6 However, the underlying causes of keloid formation remain poorly understood, and effective clinical treatments are still lacking.
At present, a large number of studies have shown that keloids are closely related to programmed cell death (PCD). As early as 1996, Ian et al found that numerous cells with the typical condensed nucleus characteristic of apoptotic cells were seen at the interface between the dermis and keloid, although none were observed in the most central part of the keloid.7 In 2001, Teofoli et al found that apoptotic cells were detected in both normal cells and keloid-derived fibroblasts, but the number of apoptotic cells in normal cells was twice that of all keloid-derived fibroblasts.8 A study by Lee et al in 2022 showed that keloid tissue is defective in autophagy, because the expression of IL-17, HIF-1a, and STAT3 was significantly increased in keloid tissue, and autophagosome-to autophagolysosome conversion was defective in KF.9 In addition, Jeon et al (2019) successfully inhibited keloid fibroblasts by inducing apoptosis using the High-Mobility Group Box 1 Protein Inhibitor (Glycyrrhizin).10 Lee et al study in 2017 showed that the application of knockdown mortalin (a mitochondrial chaperone of the heat shock protein 70 family and its pro-proliferative and anti-apoptosis functions could be associated with keloid pathogenesis) to induce apoptosis of keloid spherical cells has important significance for the treatment of keloid.11
Copper plays a dual role in biological processes. As an enzyme cofactor, it plays a vital role in various biological processes. However, excessive copper levels can directly bind to the lipoylated component of the tricarboxylic acid cycle. This leads to the accumulation of lipoylated proteins and the loss of iron-sulfur cluster proteins, resulting in proteotoxic stress and eventual cell death, known as cuproptosis.12,13 Elesclomol, a potent copper ion carrier, has been shown to induce cell death in a copper-dependent manner. Postmortem analysis of a Phase 3 combination clinical trial revealed that elesclomol exhibited antitumor activity in melanoma patients with low plasma lactate dehydrogenase (LDH) levels.14 Keloid, similarly, is a disease characterized by abnormal cell proliferation, and the lack of copper-induced cell death may contribute to keloid progression.
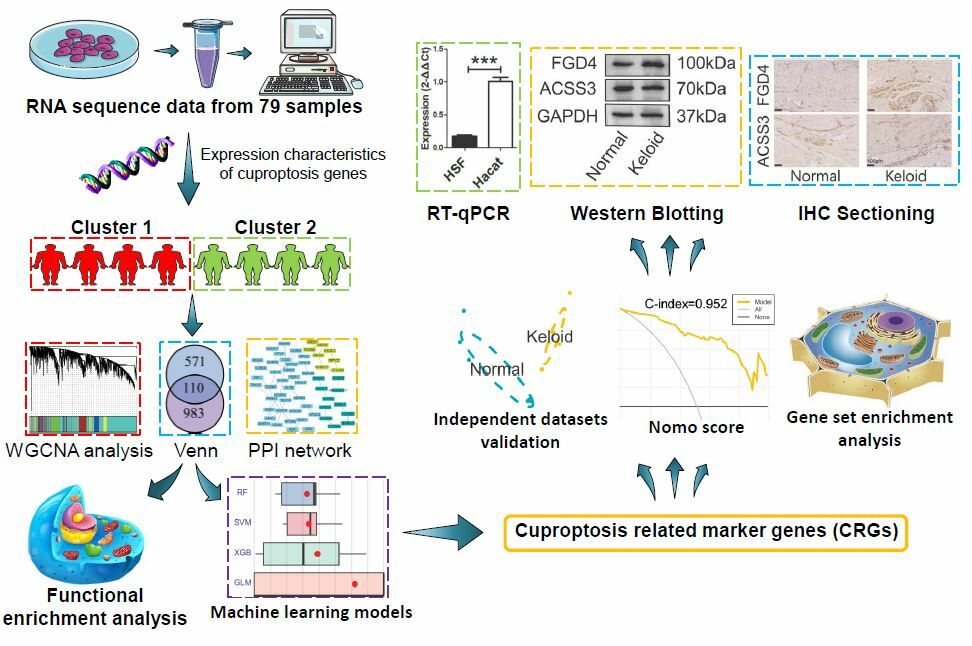
After a thorough review of the existing literature, we have identified a significant research gap regarding the association between cuproptosis and the occurrence of keloids. Therefore, the objective of this study was to investigate the involvement of cuproptosis-related genes in keloid development at the transcriptome level. Figure 1 briefly showed the process of the study.
 |
Figure 1 The experiment flowchart and analysis flowchart. |
Materials and Methods
The Datasets Source, Cell Line, and Patient Sample
In this study, we obtained five datasets from the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/). The datasets used were GSE44270 and GSE145725, which served as the training group for identifying keloid-related genes for diagnosis. The testing group consisted of GSE7890, GSE92566, and GSE121618. To address the issue of batch effects and ensure data standardization, we utilized two R packages: “sva”15 and “limma”.16
The HSF cell line (human skin fibroblasts) and Hacat cell line (human skin keratinocytes) were from the Kunming Cell Bank of Typical Culture Preservation Committee of Chinese Academy of Sciences (Kunming, China). The excised keloid sample was approved and informed consent by the Clinical Research Ethics Committee of Huizhou First Hospital and according to the Helsinki Declaration of Principles, obtained from an adult patient undergoing elective scar resection at the Huizhou First Hospital. Patient sample was used for Western blotting and immunohistochemical sectioning.
Identification of Cuproptosis-Related Candidate Genes for Keloid
In this study, we retrieved a set of 27 cuproptosis-related genes from the ferroptosis database.17 Using the expression levels of these 27 cuproptosis genes, we performed a clustering analysis on all the samples in the training group, resulting in the identification of two cuproptosis clusters. Next, we conducted weighted gene co-expression analysis (WGCNA) to identify genes that were correlated with the cuproptosis clusters. From this analysis, we selected two gene sets that exhibited the highest correlation with the cuproptosis clusters. Furthermore, we applied WGCNA to discover differentially expressed gene sets between normal samples and keloid samples. Similarly, we selected two gene sets with the highest correlation using this analysis. To identify cuproptosis-related candidate genes, we intersected the cuproptosis cluster gene sets with the keloid gene sets. This intersection allowed us to obtain a set of candidate genes that were potentially linked to both cuproptosis and keloid development. These cuproptosis-related candidate genes were further subjected to functional enrichment analysis to gain insights into their biological functions. Additionally, they were utilized for building machine learning models, potentially contributing to the development of diagnostic or predictive tools for cuproptosis and keloid-related conditions.
Functional Enrichment Analysis
To predict the potential functions of cuproptosis-related candidate genes, we conducted functional enrichment analysis to identify significantly enriched Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. The functional enrichment analysis was carried out using the clusterProfiler software in R (version 4.2.2).18 Furthermore, we utilized gene set variation analysis (GSVA) to explore the potential gene sets that might be influenced by keloid characteristic genes. The gene sets used in the GSVA analysis (C2 gene set) were obtained from the MSigDB database (http://www.gsea-msigdb.org/gsea/msigdb/). P value < 0.05 was the filter for functional enrichment analysis.
Search for Keloid Marker Genes Based Machine Learning
Based on the cuproptosis-related candidate genes, we utilized four machine learning models - Support Vector Machine (SVM), Random Forest (RF), Generalized Linear Model (GLM), and Extreme Gradient Boosting (XGB) - with the aim of identifying reliable keloid diagnostic genes.
SVM algorithm enables to generate a hyperplane in the characteristic space to distinguish between positive and negative instances with a maximum margin. Given a labeled training dataset:
where yi the class label (negative or positive) of a training compound i and xi is a feature vector representation. The optimal hyperplane can be defined as following: wxT + b=0, where x is the input feature vector, w is the weight vector, and b is the bias. Vectors xi for which |yi| (wxiT + b)= 1 will be termed support vector.
RF is an ensemble machine learning approach utilizing various independent decision trees for the prediction of classification or regression.
We used the method “rfsrc” in the R package “randomForestSRC” to construct a random forest model and selected features. The the random forest score was established by the following formula:
where Ei is the expression of feature gene i, and ri is the characteristic co-efficient of feature gene i.
The GLM is an extension of the traditional linear model, which is an algorithm in which the population mean is passed by a nonlinear join function to better process and obtain non-normally distributed data. The calculation formula for a generalized linear model can be represented as:
Here, g() is the link function, E(Y) is the expected value (mean) of the response variable Y, β0 is the intercept, and β1, β2, …, βp are the weight coefficients for the predictors x1, x2, …, xp.
XGB algorithm is a gradient boosting method which combines the regression tree. The goal function of the XGB algorithm model is:
where L(θ) is the training loss function, and Ω(θ) is the complexity function. K represents the number of trees, F represents all possible DT, and f denotes a specific CART tree. wi is the score on the ith leaf node, and T is the number of leaf nodes in the tree.
To compare the performance of these models, we evaluated them using two metrics: Root Mean Square Error (RMSE) and Receiver Operating Characteristic (ROC) curve analysis. RMSE provides a measure of the prediction error, while the ROC curve assesses the models’ ability to discriminate between keloid and non-keloid samples. By comparing the RMSE values and analyzing the ROC curves, we identified the most reliable machine learning model among the four. Using the final selected machine learning model, we determined the cuproptosis-related genes (CRGs) with the highest importance scores. Next, we defined the CRGs-based Nomo score.
The nomogram is a graphical tool used to assess the risk of a particular diagnosis. It simplifies the use of prediction models by converting the values of multiple predictor variables into a simple linear scale. A typical nomogram consists of a baseline and several vertical lines. Each vertical line represents a predictor variable and has corresponding scale values. By connecting the scale values of each predictor variable to the baseline and finding the corresponding points on the baseline, a total score can be obtained. This Nomo score can be used to evaluate the risk of a particular diagnosis. The formula of Nomo score was as follows:
Here, Nomo score is the risk score of each patient. The n is the number of gene. The coef (Genei) is the coefficient of Genei obtained by regression analysis of nomogram. Expression (Genei) is the expression level of Genei.
Validation of Characteristic Genes by Using Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
Firstly, total RNA was extracted using TRlzol reagent. Subsequently, cDNA synthesis was conducted using the StarScript III All-in-one RT Mix with gDNA Remover (A230). Next, RT-qPCR was performed utilizing 2× Fast SYBR Green Master Mix (Roche Diagnostics, Basel, Switzerland) on a LightCycler 480 (Roche Diagnostics). The primer sequences employed are presented in Table 1. Finally, the 2−ΔΔCt method was employed to evaluate the expression of the characteristic genes.
 |
Table 1 Primers for Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR) |
Western Blotting
Proteins were extracted using a protein extraction kit, and cells were lysed in the presence of protease inhibitors and phosphatase inhibitors. Protein concentrations were determined using the BCA protein kit. Subsequently, the proteins were loaded onto SDS gels and subjected to electrophoresis for 35 minutes. Following electrophoresis, the proteins were transferred onto PVDF membranes within 30 minutes. The membranes were then blocked using milk and incubated with primary antibodies overnight. After washing three times with TBST, the membranes were incubated with secondary antibodies for 60 minutes at room temperature. The protein bands were visualized using an ECL chromogenic solution.
Immunohistochemical Verification
The fresh tissue was initially fixed using 4% paraformaldehyde and subsequently embedded in paraffin following gradient dehydration. Paraffin sectioning mechanism was employed to prepare sections with a thickness of 5μm. These sections were then subjected to histochemical staining using specific antibodies to evaluate the features.
Statistical Analysis
The statistical analysis software we used was R-version 4.2.2, which helped us analyze data and create figures and tables (https://www.r-project.org). We also drew diagrams with the help of an online website called bioinformatics (https://www.bioinformatics.com.cn/).
Results
Identification of Cuproptosis-Related Candidate Genes for Keloid
The distribution of 27 cuproptosis genes on chromosomes was first shown, and they are scattered on chromosomes (Figure 2A). Subsequently, the expression levels of cuproptosis genes were compared between normal and keloid samples in the training group. Notably, five cuproptosis genes—FDX1, CISD1, DNA2, CDKN2A, and HSPA1A—exhibited differential expression between the two sample types (Figure 2B and C). Among these, FDX1 and CDKN2A were found to be upregulated in keloid samples, potentially indicating a risk factor associated with their increased expression. Conversely, CISD1, DNA2, and HSPA1A showed higher expression in normal samples, suggesting a potential protective factor associated with their upregulation. Furthermore, a correlation analysis of cuproptosis genes was conducted, revealing associations in their expression patterns within keloid tissue (Figure 2D–E). In the analysis of genes co-expression, the absolute value of correlation coefficient was greater than 0.4 in 16%, and the P-value was less than 0.05 in 17% (Figure 2E). To further investigate the heterogeneity within keloid samples, the expression profiles of cuproptosis genes were utilized to classify the training group samples into two distinct clusters, namely C1 and C2 (Figure 2F–G). This clustering approach shed light on the existence of subgroups within keloid samples based on the expression of cuproptosis genes. The reliability of the clustering method was confirmed through PCA analysis (Figure 2H), which provided additional support for the separation of keloid samples into distinct groups based on cuproptosis gene expression patterns. In addition, nine cuproptosis genes—DLD, SLC31A1, CDK5RAP1, ETFDH, HSPA1A, HSPA1B, ATP7A, ATP7B, and GLS—were found to be differentially expressed between two clusters (Figure 2I and J).
To further explore the relationship between cuproptosis and keloid formation, a Weighted Gene Co-expression Network Analysis (WGCNA) was conducted based on cuproptosis clusters. In this analysis, all genes were categorized into six gene sets based on their biological correlation (Figure S1A–C). Among these sets, the two gene sets exhibiting the highest correlation were selected as cuproptosis cluster gene sets: MEbrown (cor=0.71, P=4e−05) and MEturquoise (cor=−0.69, P=6e−05) (Figure 3D). Consequently, a total of 1093 genes were identified as cuproptosis cluster gene sets. Additionally, WGCNA analysis was performed to compare normal and keloid samples. All genes were divided into ten gene sets (Figure S1E–G). The two gene sets with the highest correlation were MEblack (cor=0.36, P=0.009) and MEbrown (cor=−0.36, P=0.009) (Figure S1H), resulting in the identification of 681 genes from MEblack and MEbrown, which were referred to as keloid gene sets. By intersecting the cuproptosis cluster gene sets and the keloid gene sets, a total of 110 cuproptosis-related candidate genes were obtained (Figure 3A).
Functional Enrichment Analysis
To further investigate the biological roles of the 110 cuproptosis-related candidate genes in keloid formation, GO and KEGG enrichment analyses were conducted. In the GO analysis, a total of 57 significant pathways were identified. These pathways encompassed 13 biological process (BP) terms, 14 cell component (CC) terms, and 30 molecular function (MF) terms. Among these, the ten BP, CC, and MF pathways with the lowest P-values were selected and presented in the diagrams (Figure 3B). The majority of the GO terms were associated with cell growth, which aligns with the dysplastic characteristics of keloid formation. This observation suggests that the cuproptosis candidate genes may be involved in the regulation of keloid growth, as indicated by their enrichment in pathways related to cell growth processes.
In the KEGG analysis, a total of nine meaningful pathways were identified. Notably, among the cuproptosis candidate genes, the MAPK signaling pathway exhibited the largest number of genes (Figure 3C). The MAPK signaling pathway is well-known for its impact on cell growth processes, which once again corresponds to the characteristics of keloid formation. This finding suggests that the MAPK signaling pathway may be dysregulated and serve as a crucial molecular basis underlying keloid development. The GO and KEGG enrichment analyses provide valuable insights into the potential biological roles of the cuproptosis-related candidate genes in keloid formation. The identified pathways, particularly those associated with cell growth and the MAPK signaling pathway, highlight potential molecular mechanisms involved in keloid pathogenesis. Further studies focused on these pathways may contribute to a better understanding of keloid development and the development of targeted therapeutic approaches. The protein-protein Interaction Network demonstrated the association between 110 cuproptosis-related candidate genes (Figure 3D).
Search for Keloid Marker Genes Based Machine Learning
To assess the predictive power of the 110 cuproptosis-related candidate genes, machine learning models were constructed using SVM, RF, GLM, and XGB algorithms. The residual distributions of the four models were calculated and compared (Figure 3E and F). Among them, the RF machine learning model exhibited the smallest root mean square of residuals (RMSE) (Figure 3E), indicating its superior performance in predicting keloid samples. The predictive reliability of the four models was further evaluated using ROC curves (Figure 3G). The SVM model achieved an Area Under the Curve (AUC) value of 0.929, while the RF model attained an AUC value of 0.857. These results suggest that the SVM and RF models have good predictive capabilities. SVM model screened 19 characteristic genes for keloid (Figure 3H). The RF model had the lowest error rate in 371 trees, and there were 17 genes with importance scores greater than 0.4 (Figure 3I and J). Through the intersection of SVM and RF characteristic genes, we obtained 15 candidate genes (Figure 3K and L). The 15 candidate genes had high expression correlation and most of them were differentially expressed between normal and keloid samples (Figure 4A and B). Ten candidate genes with AUC greater than 0.65 were used for further analysis.
In order to quantitatively evaluate the degree of danger of candidate genes, a novel Nomo score was also constructed in the training group. Among them, only the coefficients of FGD4 and ALPK2 are positive, which means that they are risk factors for keloid (Figure 4C). The specific calculation formula is as follows: Nomoscore=1.013*FGD4-4.3319*ADRB2-1.7197*KRT33B-0.163*HOXB2-1.959*RASSF7-1.4435*HAS2-3.2944*AVPI1-5.5632*ACSS3+0.5752*ALPK2-2.0643*FGFR1. The Nomo score has an extremely high degree of fit (Figure 4D-F). The key gene of Nomo score was named cuproptosis-related genes (CRGs). In the training group, the AUC value of Nomo score to diagnosis keloid was 0.927 (Figure 4G). In PCA analysis, the normal group and the keloid group were well distinguished by the expression of CRGs (Figure 4H). In the training group, the AUC value of Nomo score was 1.000 (Figure 4I). In three independent data sets of the testing group, the normal group and the keloid group were well distinguished by the expression of CRGs (Figure 4J–L). The AUC value of Nomo score in the total group was 0.952 (Figure 4M). There were also statistically significant differences in the distribution of Nomo score among different groups (Figure 4N).
Validation of Characteristic Genes by Using RT-qPCR
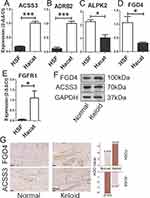
Considering that there are only 5 genes in CRGs that have been poorly studied in the field of keloid progression, we compared the expression differences of these five genes in HSF cell line and Hacat cell line by RT-qPCR. And the analysis revealed that the expression level of ALPK2 and FGD4 were higher in HSF cell line, while ACSS3, ADRB2, and FGFR1 were higher in Hacat cell line (Figure 5A–E). These were consistent with the results described above.
Protein Expression in Patient Sample
We selected two genes with the highest Nomo coefficient, one positive and one negative, and tested their corresponding protein expression levels in patient sample. In Western Blotting experiments, FGD4 was highly expressed in keloid sample, while ACSS3 was highly expressed in adjacent normal sample (Figure 5F), which was consistent with the biological characteristics shown by their Nomo coefficients. In addition, in immunohistochemical experiments, FGD4 was also highly expressed in keloid sample, while ACSS3 was also highly expressed in adjacent normal sample, which was consistent with the above results (Figure 5G). The semi-quantitative results using image J software were also consistent with this. The average optical density (AOD) value of FGD4 was higher in keloid tissues, while the AOD value of ACSS3 was higher in normal sample (Figure 5G).
Gene Set Variation Analysis of CRGs
To investigate the potential gene sets influenced by keloid characteristic genes, the GSVA analysis was performed on all keloid samples in the overall group. The analysis revealed that the high expression of CRGs was closely associated with specific gene sets, including biosynthesis of keratan sulfate and ECM receptor interaction (Figure S1I). This suggests that CRGs may play a role in these biological processes and pathways, potentially contributing to keloid formation. Conversely, the low expression of CRGs was predominantly linked to various signaling pathway gene sets, such as the nod-like receptor signaling pathway, epithelial cell signaling, and ERBB signaling pathway (Figure S1I).
Discussion
We observed differential expression of the cuproptosis gene between normal and keloid samples. Consequently, we classified all keloid samples into two clusters based on the expression levels of the cuproptosis gene. Utilizing WGCNA analysis, we identified gene sets specific to each cuproptosis cluster. Additionally, by performing WGCNA analysis comparing normal and keloid samples, we obtained gene sets specific to keloids. By intersecting these two gene sets, we identified 110 candidate genes associated with cuproptosis. Subsequent functional enrichment analysis revealed that these genes were primarily involved in cell growth and the MAPK signaling pathway. The development of keloids is closely associated with the excessive overgrowth of fibrocytes.19 And the MAPK signaling pathway is one of the most important pathways of cell proliferation.20 The functional enrichment analysis results suggest that cuproptosis-related candidate genes may play a regulatory role in cell growth within keloid tissue through the MAPK pathway. To predict keloid characteristic genes, we comprehensively evaluated the performance of four machine learning models and identified 10 genes as CRGs. The Nomo score was constructed based on CRGs. Then we selected two genes with the highest Nomo coefficient (FGD4 and ACSS3) and tested their corresponding protein expression levels in patient sample. The results of Western blotting and IHC experiments were consistent with the above conclusions.
Furthermore, the GSVA analysis indicated that the elevated expression of CRGs was closely associated with gene sets related to the biosynthesis of keratan sulfate and ECM receptor interaction. The ECM has been consistently identified as a significant contributor to fibrosis, including keloid formation. This connection between CRGs and ECM receptor gene sets suggests that the ECM may play a crucial role in the characteristic gene profile of keloids.21,22 The results of GSVA analysis showed that there was a relationship between CRGs and ECM receptor gene set, which may be an important reason why CRGs was a characteristic gene, but the specific mechanism needs to be further studied.
In conclusion, while we have identified characteristic genes, predicted keloid-related signaling pathways, and suggested potential therapeutic targets, it is important to note that all data sets used in this study were retrospective. Therefore, prospective analysis is necessary to validate our findings. In any case, we plan to address these questions in further studies, taking a step-by-step approach to better understand the underlying mechanisms of keloid formation and progression.
Conclusion
In this study, we utilized the five datasets to identify genes associated with keloid diagnosis. The WGCNA analysis found candidate genes related to different cuproptosis clusters. Functional enrichment analysis revealed that these candidate genes were significantly associated with cell growth and the MAPK signaling pathway. We integrated four machine learning models and identified CRGs for keloid diagnosis. The reliable diagnostic capabilities of CRGs were demonstrated through three independent datasets, RT-qPCR, Western blotting, and IHC section.
Data and Code Availability
The data and code used for analyses in this study can be found at https://github.com/wxmm20230126/Computational-Framework/blob/main/Public%20code_keloid.R.
Ethical Approval and Consent to Participate
The work was approved by the Clinical Research Ethics Committee of Huizhou First Hospital. Informed consent forms are not required for patient data extracted from public databases.
Acknowledgments
We are grateful to everyone who provides and builds public data.
Informed Consent Statement
Informed consent was received from each patient participating in the study.
Consent for Publication
All authors gave consent to publish.
Disclosure
The authors declare that there are no conflicts of interest regarding the publication of this study.
References
1. Wynn TA, Ramalingam TR. Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat Med. 2012;18(7):1028–1040. doi:10.1038/nm.2807
2. Rinkevich Y, Walmsley GG, Hu MS, et al. Skin fibrosis. Identification and isolation of a dermal lineage with intrinsic fibrogenic potential. Science. 2015;348(6232):aaa2151. doi:10.1126/science.aaa2151
3. Andrews JP, Marttala J, Macarak E, et al. Keloids: the paradigm of skin fibrosis - Pathomechanisms and treatment. Matrix Biol. 2016;51:37–46. doi:10.1016/j.matbio.2016.01.013
4. Murota H, Lingli Y, Katayama I. Periostin in the pathogenesis of skin diseases. Cell Mol Life Sci. 2017;74(23):4321–4328. doi:10.1007/s00018-017-2647-1
5. Do NN, Eming SA. Skin fibrosis: models and mechanisms. Curr Res Transl Med. 2016;64(4):185–193. doi:10.1016/j.retram.2016.06.003
6. Mustoe TA. Scars and keloids. BMJ. 2004;328(7452):1329–1330. doi:10.1136/bmj.328.7452.1329
7. Appleton I, Brown NJ, Willoughby DA. Apoptosis, necrosis, and proliferation: possible implications in the etiology of keloids. Am J Pathol. 1996;149(5):1441–1447.
8. Luo S, Benathan M, Raffoul W, et al. Abnormal balance between proliferation and apoptotic cell death in fibroblasts derived from keloid lesions. Plast Reconstr Surg. 2001;107(1):87–96. doi:10.1097/00006534-200101000-00014
9. Lee SY, Lee AR, Choi JW, et al. IL-17 Induces Autophagy Dysfunction to Promote Inflammatory Cell Death and Fibrosis in Keloid Fibroblasts via the STAT3 and HIF-1alpha Dependent Signaling Pathways. Front Immunol. 2022;13:888719. doi:10.3389/fimmu.2022.888719
10. Jeon YR, Roh H, Jung JH, et al. Antifibrotic Effects of High-Mobility Group Box 1 Protein Inhibitor (Glycyrrhizin) on Keloid Fibroblasts and Keloid Spheroids through Reduction of Autophagy and Induction of Apoptosis. Int J Mol Sci. 2019;20(17):4134. doi:10.3390/ijms20174134
11. Lee WJ, Ahn HM, Na Y, et al. Mortalin deficiency suppresses fibrosis and induces apoptosis in keloid spheroids. Sci Rep. 2017;7(1):12957. doi:10.1038/s41598-017-13485-y
12. Tsvetkov P, Coy S, Petrova B, et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science. 2022;375(6586):1254–1261. doi:10.1126/science.abf0529
13. Cobine PA, Brady DC. Cuproptosis: cellular and molecular mechanisms underlying copper-induced cell death. Mol Cell. 2022;82(10):1786–1787. doi:10.1016/j.molcel.2022.05.001
14. O’Day SJ, Eggermont AMM, Chiarion-Sileni V, et al. Final results of Phase III SYMMETRY study: randomized, double-blind trial of elesclomol plus paclitaxel versus paclitaxel alone as treatment for chemotherapy-naive patients with advanced melanoma. J Clin Oncol. 2013;31(9):1211–1218. doi:10.1200/JCO.2012.44.5585
15. Leek JT, Johnson WE, Parker HS, et al. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28(6):882–883. doi:10.1093/bioinformatics/bts034
16. Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi:10.1093/nar/gkv007
17. Zhou N, Yuan X, Du Q, et al. FerrDb V2: update of the manually curated database of ferroptosis regulators and ferroptosis-disease associations. Nucleic Acids Res. 2023;51(D1):D571–D582. doi:10.1093/nar/gkac935
18. Yu G, Wang L-G, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi:10.1089/omi.2011.0118
19. Trace AP, Enos CW, Mantel A, et al. Keloids and Hypertrophic Scars: a Spectrum of Clinical Challenges. Am J Clin Dermatol. 2016;17(3):201–223. doi:10.1007/s40257-016-0175-7
20. Fang JY, Richardson BC. The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 2005;6(5):322–327. doi:10.1016/S1470-2045(05)70168-6
21. Deng -C-C, Hu Y-F, Zhu D-H, et al. Single-cell RNA-seq reveals fibroblast heterogeneity and increased mesenchymal fibroblasts in human fibrotic skin diseases. Nat Commun. 2021;12(1):3709. doi:10.1038/s41467-021-24110-y
22. Griffin MF, desJardins-Park HE, Mascharak S, et al. Understanding the impact of fibroblast heterogeneity on skin fibrosis. Dis Model Mech. 2020;13(6). doi:10.1242/dmm.044164
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.