Back to Journals » Cancer Management and Research » Volume 10
Differences in clinical characteristics and mutational pattern between synchronous and metachronous colorectal liver metastases
Authors Zheng P, Ren L, Feng Q, Zhu D, Chang W , He G, Ji M, Jian M , Lin Q, Yi T, Wei Y , Xu J
Received 3 January 2018
Accepted for publication 12 April 2018
Published 27 August 2018 Volume 2018:10 Pages 2871—2881
DOI https://doi.org/10.2147/CMAR.S161392
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Leylah Drusbosky
Peng Zheng,* Li Ren,* Qingyang Feng,* Dexiang Zhu,* Wenju Chang, Guodong He, Meiling Ji, Mi Jian, Qi Lin, Tuo Yi, Ye Wei, Jianmin Xu
Department of General Surgery, Zhongshan Hospital, Fudan University, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Purpose: To investigate differences in clinical characteristics and mutational patterns between synchronous and metachronous colorectal liver metastases (CLMs).
Patients and methods: From June 2008 to December 2014, patients with RAS wild-type CLMs treated at Zhongshan Hospital, Fudan University were included. DNA extracted from formalin-fixed paraffin-embedded tissue of primary tumors was sequenced with next-generation sequencing for single-nucleotide polymorphism of 96 genes according to custom panel. Mutations were compared between synchronous and metachronous liver metastases and correlated with clinical characteristics.
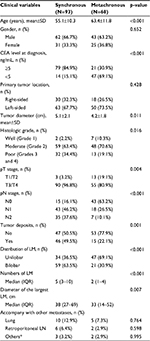
Results: A total of 161 patients were included: 93 patients with synchronous CLM and 68 patients with metachronous CLM. Patients with metachronous CLM were obviously elder. For pathology of primary tumors, synchronous CLMs were larger in size, poorly differentiated, and more frequently local advanced and lymph node positive. For evaluation of liver metastases, synchronous CLM had more and larger metastatic lesions. The median number of mutations in synchronous CLMs was significantly higher than in metachronous group (22 vs. 18, p<0.001). EGFR rs2227983 is the most prevalent mutation in both groups and only a part of prevalent mutations is shared in both groups. Prevalent mutations were correlated with many clinical characteristics. EGFR rs2227983, RBMXL3 rs12399211, and PTCH1 rs357564 were prognostic for latency of metachronous CLM.
Conclusion: Clinically, synchronous CLMs, compared with metachronous CLMs, were younger and showed heavier tumor burden for both primaries and liver metastases. Genetically, we identified different mutational patterns between synchronous and metachronous CLMs and several correlations between mutations and clinical characteristics. Further researches were needed to confirm these potential key mutations of CLMs.
Keywords: colorectal liver metastases, next-generation sequencing, metachronous, synchronous
Introduction
Colorectal cancer (CRC) is the most common malignancy throughout the world,1 and liver metastases are the major cause of death in CRC patients. Unfortunately, 40%–50% of CRC patients would develop colorectal liver metastases (CLMs). They are either synchronous or metachronous in presentation with approximately equal incidence.2,3 Compared with synchronous metastases, patients with metachronous metastases show differences in terms of clinicopathologic characteristics and better prognosis after metastases resections.4–6 However, in previous studies, synchronous and metachronous liver metastases were usually lumped together, neglecting their clinical and biologic differences. It is critical to improve our understanding of the biology of liver metastases, which may help to develop more effective therapeutic strategies.
Biologic differences between synchronous and metachronous CLMs had been studied previously but showed conflicting results.2,7–11 Available evidence indicated that majority of gene alterations in the primary tumor were maintained in the CLMs, whereas a limited number of studies compared the primary tumors of synchronous and metachronous groups and generally demonstrated no differences in biomarker expression. In consideration of limited number of studies and limited number of biomarkers investigated, it is still not reasonable to deny the hypothesis that the biology of synchronous and metachronous CLMs would be different in view of the known clinicopathologic differences that exist between them. Thus, more comprehensive researches are needed. The use of next-generation sequencing (NGS) for high-throughput genomic analysis has accelerated our understanding of the molecular characteristics of CRC12–14 and may also serve us an insight on this topic.
Therefore, we conducted NGS of primary tumors of synchronous or metachronous CLM to investigate mutational patterns between them. We also expected to find prognostic or predictive biomarkers among mutations sequenced.
Patients and methods
Study population
This study retrospectively included patients with CRC treated at General Surgery Department of Zhongshan Hospital, Fudan University (Shanghai, China) during June 2008 to December 2014. The inclusion criteria were as follows: colorectal adenocarcinoma determined by pathologic evidence; wild-type RAS; liver metastases determined by radiologic and/or pathologic evidence; radical resections of primary tumors; enough formalin-fixed paraffin-embedded (FFPE) tissue of primary tumors; no exposure to any treatment (chemotherapy, targeted therapy, radiotherapy, or interventional therapy) before primary tumor resections. Patients with liver metastases accompanied with other distant metastases were permitted. Adjuvant chemotherapy after primary tumor resection was permitted. Target therapy and interventional therapy were permitted only after the occurrence of liver metastases. All patients provided written and oral informed consent. This study was approved by the ethics committee of Zhongshan Hospital, Fudan University.
Two groups were established in this study: the synchronous-metastases group and the metachronous-metastases group. The synchronous-metastases group consisted of patients with liver metastases diagnosed together with or within a 6-month interval of the diagnosis of the primary colorectal tumor. The metachronous-metastases group consisted of patients with liver metastases diagnosed >6 months after primary tumor resection. This study was approved by the local ethics committees and all patients provided written and oral informed consent, including research on tumor tissue.
In addition, 10 patients were selected for whole-exome sequencing (WES). All 10 patients were diagnosed with CLM and underwent resection of both primary and metastatic tumors. Exclusion criteria included previous exposure to any treatment for metastatic CRC, family history of CRC, and evidence of a mismatch repair deficiency.
Study procedure
To investigate mutational pattern of different liver metastases, we examined biomarkers (single-nucleotide polymorphism, SNP) through genome-wide exploration using NGS. To preliminarily select genes to construct a custom panel for target capture sequencing, we performed WES for 10 triplets, each comprising primary colorectal tumor and normal colorectal mucosa and matched liver metastases. Genomic DNA from fresh tissue was samples sequenced on an Ion™ Proton (Life Technologies, Carlsbad, CA, USA) platform according to the manufacturer’s instructions. Normal colorectal mucosa was sequenced to exclude germline variants. The read alignments and variant analyses were performed according to the predefined workflow.
We constructed a custom panel of 96 genes selected based on driver mutations identified using WES and Tumor Mutation Hotspots Panel version 2 (Life Technologies, Carlsbad, CA, USA). Genomic DNA from FFPE tissue samples of patients in both cohorts was subsequently sequenced for SNPs using an Ion™ Torrent Personal Genome Machine (PGM) according to the manufacturer’s instructions. For a given gene loci, the fraction of mutant alleles was calculated by diving the number of mutant reads by the number of total reads. A 5% cutoff value was employed. A sample was considered wild-type for a given gene when all sequenced loci harbored <5% mutant alleles.
Whole-exome sequencing
DNA was extracted from fresh tumor samples using the MELT™ Total Nucleic Acid Isolation Kit (Life Technologies). Quantity and quality were assessed using Qubit 2.0 (Life Technologies). Fifty to hundred nanograms of DNA for each sample was used for exome capture and library preparation with Ion AmpliSeq™ Exome Kit 4xDuo (Life Technologies) following the manufacturer’s instructions. Then libraries were bar-coded with Ion Xpress™ Barcode Adapters Kit (Life Technologies). The concentration of each library was determined by PCR with the Ion Library™ Quantization Kit (Life Technologies). According to the manufacturer’s instructions, all libraries were diluted to 100 pM working solutions and then pooled as needed to perform the template preparation with Ion PI™ Template OT2 200 kit v2 (Life Technologies) on Ion One Touch™ 2 System. Quality and quantity were determined with Qubit Ion Sphere™ Quality Control Kit (Life Technologies) for the obtained ion sphere particles. WES was performed on Ion Proton™ platform, using the Ion PI™ Sequencing 200 kit v2 and Ion PI™ Chip kit v2. Sequencing data were analyzed with the Torrent Suite™ Software v4.0 (Life Technologies) using default parameters setting.
Processing of WES data
The primary WES data were analyzed for single-nucleotide variants (SNVs) following the procedures indicated below.
- Variants with SNV quality (QUAL) ≤20 were excluded. QUAL was calculated using Torrent Suite™ software.
- Variants of normal mucosa were considered background variants. Primary or metastatic tumor samples were filtered using background variants, and the variants were rejected as germline variants or sequencing artifacts when present in the corresponding normal samples.
- Primary or metastatic tumor-specific SNVs were analyzed using the SeattleSeq SNP Annotation.15 Known germline mutations from the Exome Sequencing Project16 and dbSNP databases (build 140)17 were also excluded. We selected nonsilent mutations, including missense mutations and InDels.
- Nonsilent mutations were predicted to affect gene function when any of the following criteria were fulfilled: 1) functional impact score of SIFT18 ≤0.05;19 2) functional impact score of PolyPhen-220 >0.45;21 3) functional impact label of Mutation Assessor22 was “medium” or “high”;23 4) Condel24 label was “deleterious”;25 and 5) functional impact score of FATHMM26 <0.27
- The transFIC analysis was performed as previously described.28 Mutations were considered cancer driver mutations when the outcome of the transFIC analysis was of “high impact”.
Construction of custom panel
To prepare for target capture sequencing, we constructed a custom panel based on WES data and Tumor Mutation Hotspots Panel version 2. When selecting genes from WES data, genes that were essential for cancer progression, particularly liver metastasis, were considered with priority. On one hand, we searched all mutations in the GeneRIF database29 using key words “cancer/tumor/carcinoma” and “metasta-/invasion/invade/invasive/migrate”. On the other hand, we focused on mutations of universal genes in primary tumors and corresponding liver metastases. In addition, most genes in the Tumor Mutation Hotspots Panel version 2 were included. The mutation information for these genes was acquired from the Catalog of Somatic Mutations in Cancer (COSMIC) database,30 and we employed the most frequent mutations to build the panel.
Target capture sequencing
DNA was extracted from FFPE tumor samples using the RecoverAll™ Total Nucleic Acid Isolation Kit (Life Technologies) according to the manufacturer’s instructions. Quantity and quality were assessed using Qubit 2.0 (Life Technologies). Ten nanograms of DNA for each sample was used for library construction and template preparation with same procedures described above in the “Whole-exome sequencing” section. Target capture sequencing was carried out with customized panel using the Ion PGM™ platform (Life Technologies) according to the manufacturer’s instructions. The panel consisted of two separate PCR primer pools covering recurrent mutations in 96 genes with 1500X sequence coverage on Ion™ 318 chip. Sequencing data were analyzed with Ion Reporter™ software v4.4 (Life Technologies) using default parameters setting.
Statistical analysis
The statistical analysis plan was established before the genotyping results were available. Differences in categorical parameters were calculated using a chi-square test or Fisher’s exact test. Survival curves were generated using the Kaplan–Meier method and compared using a log-rank test. Hazard ratios and 95% CIs were calculated using the Cox proportional hazards model. For univariate and multivariate analyses of latency of metachronous liver metastases, the Cox proportional hazards model was used. All statistical analyses were conducted using the statistical software SPSS version 18.0 (SPSS Inc., Chicago, IL, USA). A p-value <0.05 was considered statistically significant.
Results
Patients and clinical characteristics
A total of 161 patients were included and sequenced. According to the metastatic type, 93 patients were in synchronous-metastases group and 68 patients in metachronous-metastases group. Several differences in clinical characteristics were observed between groups (Table 1). Patients with metachronous CLMs were obviously elder. With regard to pathology of primary tumors, synchronous CLMs were larger in size, poorly differentiated, and more frequently local advanced and lymph node positive. As to evaluation of liver metastases, synchronous CLMs result in more and larger metastatic lesions.
Summary of WES and target capture sequence
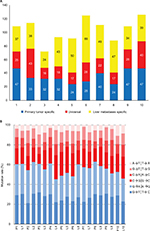
Ten triplets were sequenced with WES for preliminarily data to construct custom panel. Overall, we identified 608 potential somatic driver mutations in 511 genes in primary colorectal tumors and 694 potential somatic driver mutations in 638 genes in liver metastases. The number of somatic mutations in 10 primary colorectal tumor ranged from 41 to 87, with a mean of 60 (Figure 1A), which was not significantly different from that of the non-hypermutated CRCs reported in The Cancer Genome Atlas.31 When comparing mutations between tumors, 230 mutations in 219 genes were universal in the primary tumor and corresponding liver metastases. In addition, mutations observed in 10 patients were predominated by the C/G>T/A transition (Figure 1B) consistent with the results of previous CRC genomics studies.31,32
According to the custom panel, 161 patients were sequenced with target capture sequence for recurrent mutations of 96 genes. Mutations with mutational frequencies >10% and <90% were further investigated for stability. Finally, 48 mutations in 27 genes were included in following analysis (Table 2). Among these mutations, 40 were already documented in dbSNP databases (build 140).
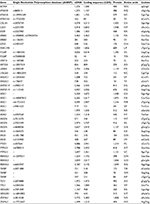  | Table 2 List of mutations analyzed |
Correlation between clinical characteristics and mutations
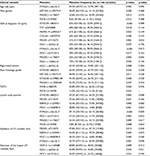
Univariate analysis identified that 18 mutations in 12 genes were correlated with clinical variables (Table 3). MDC1 was associated with the most number of clinical variables, including T stage, N stage, tumor location of primary, and size and number of liver metastases. Mutations of ATAD3B, MAGEC1, and MICB were only associated with pathologic variables of primary tumor. Most of the mutations associated with variables of liver metastases were also associated with lymph node metastases. Ten correlations with preoperative CEA level, lymph node metastases, and number of LMs remained significant with false discovery rate <0.05.
Mutational pattern of synchronous and metachronous liver metastases
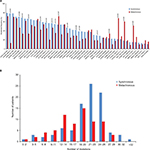
The median number of mutations in the synchronous group was 22 (range from 2 to 32), which was significantly higher than 18 (range from 2 to 28) in the metachronous group (p<0.001). Mutation frequency of the synchronous group, compared with the metachronous group, was significantly higher in 12 mutations and lower in five mutations (Figure 2). For most other mutations, the frequency was numerically higher in the synchronous group. In addition, only half of the most prevalent mutations in the synchronous group were in common with that in the metachronous group (Table 4).
  | Table 4 Prevalent mutations in synchronous and metachronous groups Notes: *p<0.05; **p<0.001. |
Mutations and survival of metachronous liver metastases
The latency of metachronous CLMs is summarized in Figure 3. Half of all metachronous CLMs occurred within 15 months, and 75% occurred within 24 months. For patients with metachronous CLMs, the latency was significantly associated with prevalent mutations in metachronous CLMs, including EGFR rs2227983, RBMXL3 rs12399211, TAPBP rs2071888, PTPN23 c.3361A>T, PTCH1 rs357564, and MDC1 rs61733213. In multivariate analysis, correlation between latency and EGFR rs2227983, RBMXL3 rs12399211, and PTCH1 rs357564 remained significant (Table 5).
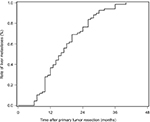  | Figure 3 Timing of metachronous metastases. Note: Half of all metachronous colorectal liver metastases occurred within 15 months, and 75% occurred within 24 months. |
  | Table 5 Univariate and multivariate analyses of latency of metachronous colorectal liver metastases Abbreviation: HR, hazard ratio. |
Discussion
In this study, we conducted NGS in primary tumors of synchronous or metachronous CLM and identified several differences in terms of clinical characteristics and mutational patterns.
For clinicopathologic characteristics, our results were broadly consistent with previous studies.2,5 Patients from the synchronous group were younger than those from the metachronous group. Compared to the metachronous group, patients in the synchronous group showed heavier tumor burden for both primaries and liver metastases. Furthermore, the synchronous group, more frequently characterized by adverse prognostic factors, had inferior overall survival than the metachronous group (median time 26 vs. 36 months, p=0.001). These results indicated that synchronous CLM may represent a more aggressive tumor subtype than metachronous CLM. All these issues highlight the need for understanding of differences in molecular biology, which results in and keeps the different manifestations between synchronous and metachronous CLMs.
Based on the understanding of the mechanisms of metastases, we extracted our efforts to identify biologic differences between synchronous and metachronous CLMs. In contrast to previous studies focusing on limited gene alterations,2,8,9,33 we used a multivariable approach, including genome-wide exploration using WES, panel construction based on bioinformatics, and selection of significant biomarkers according to clinical outcomes. The advantage of this multivariable approach is that it is unbiased by biologic assumptions and thereby may find correlations between distinct gene mutations and liver metastases.
Finally, we included and analyzed 48 mutations in 27 genes. The synchronous group harbored significantly more mutations than the metachronous group, representing a more heterogeneous and advanced subgroup of CLM. For the synchronous group, about half of the prevalent mutations were in common with the metachronous group, and the most prevalent was EGFR rs2227983. These mutations were potential key mutations of liver metastases and further research was needed. Another half of the prevalent mutations were private in the synchronous group and were significantly more frequently mutant than the metachronous group. In addition, prevalent mutations private in the metachronous group showed similar trend. Furthermore, these mutations, including RBMXL3 rs12399211, PTPN23 c.3361A>T, PTCH1 rs357564, and MDC1 rs61733213, partly explained the latency of metachronous liver metastases. All these results indicated that the synchronous and the metachronous groups showed different mutational patterns, and these mutations were correlated with several clinical characteristics. Different mutations may be potential key mutations and further researches were needed.
In previous studies on this topic, the majority found differences in molecular marker expression between CLMs and their respective primaries in both the synchronous and metachronous groups. Limited comparison was done between LMs or primaries between the synchronous and the metachronous groups. Pantaleo et al8 compared the expression signature with reverse transcriptase PCR and enzyme-linked immunosorbent assay and indicated that EGFR and COX-2 are overexpressed in metachronous and synchronous metastases, respectively. van der Wal et al2 reported that liver parenchyma adjacent to the LMs provides a highly prosperous angiogenic environment in the synchronous group compared to the metachronous groups. However, there were also many studies with negative findings.34,35 Our results added evidences for the hypothesis that synchronous and metachronous CLMs were biologically different.
According to previous studies, both synchronous and metachronous CLMs would evolve different biologic alterations to their corresponding primaries.11,32,33,36 Thus, examination of metastatic tissue is crucial as it could help choose optimal treatment according to current biologic status. Simultaneously, studies also demonstrated that the majority of biologic alterations, especially key alterations, in primaries were maintained in CLMs.11,32,35 If these key alterations of liver metastases could be detected in primaries, it could have important clinical implications for prediction of occurrence, even timing of liver metastases. Under consideration of the evidence mentioned above, we selected and examined tissue of primary tumors. If only corresponding metastatic tissue were examined, our study may provide more information.
This was a retrospective exploratory study and there were several limitations: first, we sequenced single type of gene alteration in a highly selected cohort. Only SNPs of primary tumors with wild-type RAS were sequenced. Thus, this study provided an incomplete and partial view on differences between different CLMs; second, many correlations were observed between clinical parameters and mutations, but relevant biologic evidences were still limited. More studies were needed to confirm the clinical meanings and explore the underlying mechanism. Third, the influences of tumor heterogeneity and differences between LMs and their respective primaries were not taken into consideration in the study.
Conclusion
Synchronous CLMs, compared to metachronous CLMs, were younger and showed heavier tumor burden for both primaries and liver metastases. NGS identified different mutational patterns between synchronous and metachronous CLMs. Further researches were needed to confirm the correlation between differences in clinical characteristics and mutational patterns and the mechanism inside.
Acknowledgment
This work was supported by National Natural Science Foundation of China (grant number: 81472228).
Disclosure
The authors report no conflicts of interest in this work.
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. | ||
van der Wal GE, Gouw AS, Kamps JA, et al. Angiogenesis in synchronous and metachronous colorectal liver metastases: the liver as a permissive soil. Ann Surg. 2012;255(1):86–94. | ||
Van Cutsem E, Nordlinger B, Adam R, et al; European Colorectal Metastases Treatment Group. Towards a pan-European consensus on the treatment of patients with colorectal liver metastases. Eur J Cancer. 2006;42(14):2212–2221. | ||
de Jong MC, Pulitano C, Ribero D, et al. Rates and patterns of recurrence following curative intent surgery for colorectal liver metastasis: an international multi-institutional analysis of 1669 patients. Ann Surg. 2009;250(3):440–448. | ||
Tsai MS, Su YH, Ho MC, et al. Clinicopathological features and prognosis in resectable synchronous and metachronous colorectal liver metastasis. Ann Surg Oncol. 2007;14(2):786–794. | ||
Dexiang Z, Li R, Ye W, et al. Outcome of patients with colorectal liver metastasis: analysis of 1,613 consecutive cases. Ann Surg Oncol. 2012;19(9):2860–2868. | ||
Watanabe T, Kobunai T, Yamamoto Y, et al. Prediction of liver metastasis after colorectal cancer using reverse transcription-polymerase chain reaction analysis of 10 genes. Eur J Cancer. 2010;46(11):2119–2126. | ||
Pantaleo MA, Astolfi A, Nannini M, et al. Gene expression profiling of liver metastases from colorectal cancer as potential basis for treatment choice. Br J Cancer. 2008;99(10):1729–1734. | ||
Chen J, Li Q, Wang C, Wu J, Zhao G. Prognostic significance of c-erbB-2 and vascular endothelial growth factor in colorectal liver metastases. Ann Surg Oncol. 2010;17(6):1555–1563. | ||
Nanashima A, Shibata K, Nakayama T, et al. Clinical significance of microvessel count in patients with metastatic liver cancer originating from colorectal carcinoma. Ann Surg Oncol. 2009;16(8):2130–2137. | ||
Slesser AA, Georgiou P, Brown G, Mudan S, Goldin R, Tekkis P. The tumour biology of synchronous and metachronous colorectal liver metastases: a systematic review. Clin Exp Metastasis. 2013;30(4):457–470. | ||
Yu J, Wu WK, Li X, et al. Novel recurrently mutated genes and a prognostic mutation signature in colorectal cancer. Gut. 2015;64(4):636–645. | ||
Helman E, Lawrence MS, Stewart C, Sougnez C, Getz G, Meyerson M. Somatic retrotransposition in human cancer revealed by whole-genome and exome sequencing. Genome Res. 2014;24(7):1053–1063. | ||
Sadanandam A, Lyssiotis CA, Homicsko K, et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19(5):619–625. | ||
The SeattleSeq Annotation server. Available from: http://snp.gs.washington.edu/SeattleSeqAnnotation138/. Accessed September, 2014. | ||
NHLBI Exome Sequencing Project: Exome Variant Server. Available from: http://evs.gs.washington.edu/EVS/. Accessed September, 2014. | ||
dbSNP databases. Available from: http://www.ncbi.nlm.nih.gov/projects/SNP/. Accessed September, 2014. | ||
Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1081. | ||
SIFT database. Available from: http://www.jcvi.org/. Accessed September, 2014. | ||
Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013; Chapter 7: Unit 7.20. | ||
PolyPhen-2 (Polymorphism Phenotyping v2). Available from: http://genetics.bwh.harvard.edu/pph2/. Accessed September, 2014. | ||
Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39(17):e118. | ||
Mutationassessor.org (release 3). Available from: http://mutationassessor.org/r3/. Accessed September, 2014. | ||
Gonzalez-Perez A, Lopez-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am J Hum Genet. 2011;88(4):440–449. | ||
FannsDB database: Condel. Available from: http://bbglab.irbbarcelona.org/fannsdb/. Accessed September, 2014. | ||
Shihab HA, Gough J, Cooper DN, et al. Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Hum Mutat. 2013;34(1):57–65. | ||
Fathmm (Functional Analysis through Hidden Markov Models v2.3). Available from: http://fathmm.biocompute.org.uk/. Accessed September, 2014. | ||
Gonzalez-Perez A, Deu-Pons J, Lopez-Bigas N. Improving the prediction of the functional impact of cancer mutations by baseline tolerance transformation. Genome Med. 2012;4(11):89. | ||
GeneRIF (Gene Reference Into Function). Available from: http://www.ncbi.nlm.nih.gov/gene/submit-generif. Accessed November, 2014. | ||
COSMIC (Catalog of Somatic Mutations in Cancer). Available from: https://cancer.sanger.ac.uk/cosmic. Accessed November, 2014. | ||
Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. | ||
Kim TM, Jung SH, An CH, et al. Subclonal genomic architectures of primary and metastatic colorectal cancer based on intratumoral genetic heterogeneity. Clin Cancer Res. 2015;21(19):4461–4472. | ||
Kuramochi H, Nakajima G, Kaneko Y, et al. Amphiregulin and epiregulin mRNA expression in primary colorectal cancer and corresponding liver metastases. BMC Cancer. 2012;12:88. | ||
Nakamoto RH, Uetake H, Iida S, et al. Correlations between cyclooxygenase-2 expression and angiogenic factors in primary tumors and liver metastases in colorectal cancer. Jpn J Clin Oncol. 2007;37(9):679–685. | ||
Kim KP, Kim JE, Hong YS, et al. Paired primary and metastatic tumor analysis of somatic mutations in synchronous and metachronous colorectal cancer. Cancer Res Treat. 2017;49(1):161–167. | ||
Bird NC, Mangnall D, Majeed AW. Biology of colorectal liver metastases: a review. J Surg Oncol. 2006;94(1):68–80. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.